Evaluation of the Antiviral Characteristics of Tetragonula sapiens Propolis from Indonesia Using In Vitro and In Silico Methods
Corresponding email: sahlan@eng.ui.ac.id
Published at : 25 Mar 2025
Volume : IJtech
Vol 16, No 2 (2025)
DOI : https://doi.org/10.14716/ijtech.v16i2.7192
Asih, SC, Nasikin, M, Horng, J-T, Tsai, H-P, Tsai, S-K, Chiu, C-H, Pratami, DK, Mun’im, A & Sahlan, M 2025, ‘Evaluation of the antiviral characteristics of tetragonula sapiens propolis from indonesia using in vitro and in silico methods’, International Journal of Technology, vol. 16, no. 2, pp. 686-705
| Safira Candra Asih | 1. Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia. 2. Research Center for Biomedical Engineering (RCBE), Institute of Biosystem |
| Mohammad Nasikin | Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia |
| Jim-Tong Horng | 1. Molecular Infectious Disease Research Center, Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Taoyuan 333, Taiwan. 2. Research Center for Emerging Viral Infections, Colle |
| Hui-Ping Tsai | Institute of Preventive Medicine, National Defense Medical Center, Taipei 11217, Taiwan |
| Shan-Ko Tsai | 1. Institute of Preventive Medicine, National Defense Medical Center, Taipei 11217, Taiwan. 2. Graduate Institute of Medical Science, National Defense Medical Center, Taipei, Taiwan |
| Cheng-Hsun Chiu | Molecular Infectious Disease Research Center, Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Taoyuan 333, Taiwan |
| Diah Kartika Pratami | Center for Study of Natural Product for Degenerative Disease, Faculty of Pharmacy, Pancasila University, South Jakarta, DKI Jakarta, 12640, Indonesia |
| Abdul Mun’im | National Metabolomics Collaborative Research Center, Faculty of Pharmacy, Universitas Indonesia, Kampus UI Depok, West Java, 16424, Indonesia |
| Muhamad Sahlan | 1. Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia. 2. Research Center for Biomedical Engineering (RCBE), Institute of Biosystem |
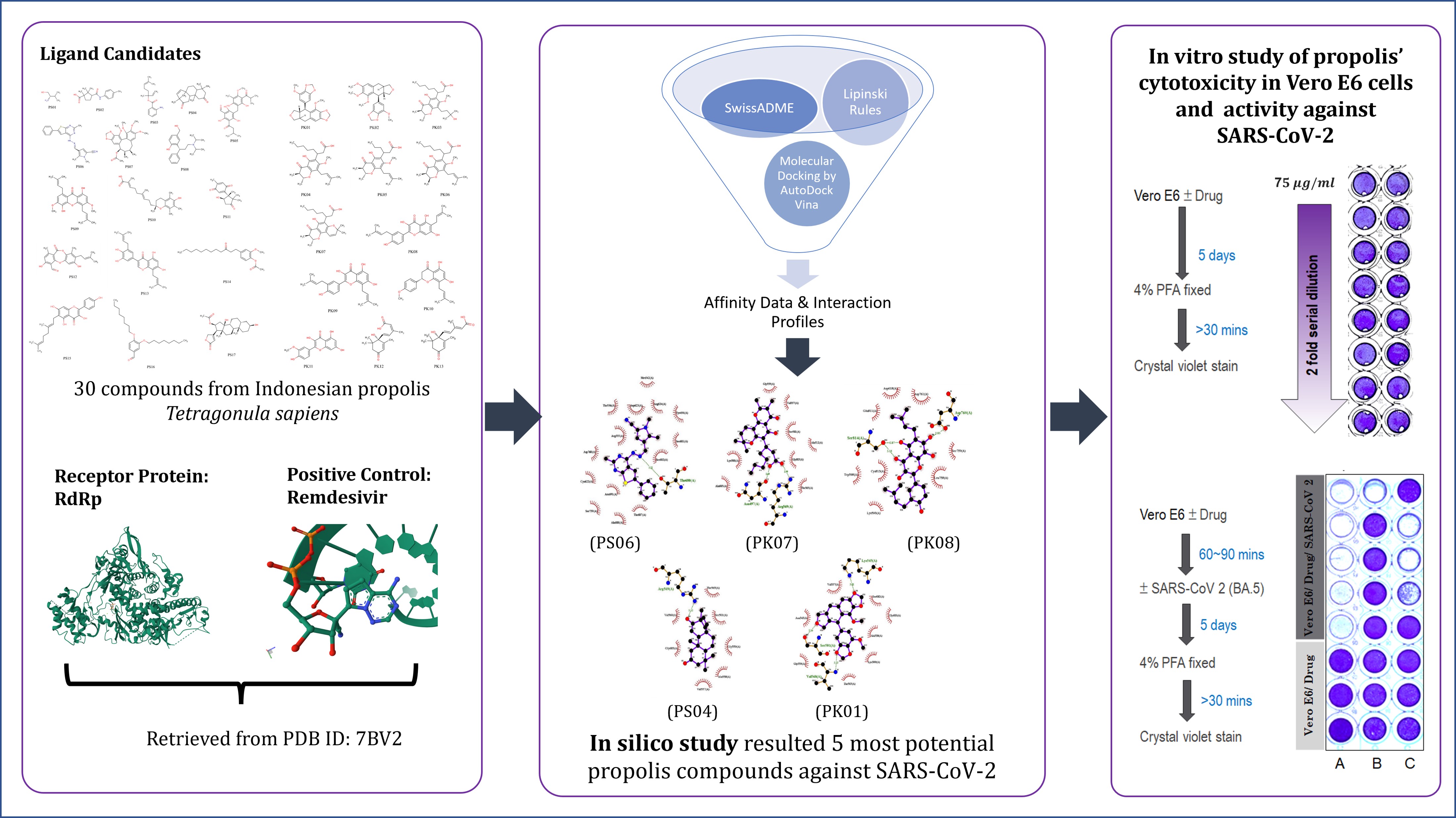
The previous outbreaks of syndrome coronavirus 2 (SARS-CoV-2) are indications of the need for effective antiviral agents to mitigate disease severity and future outbreaks. In this context, natural products such as propolis have gained attention for their antiviral properties, particularly due to the rich composition of bioactive compounds. Propolis derived from Tetragonula sapiens, a stingless bee species, has shown promising pharmacological properties, including antimicrobial, antioxidant, and anti-inflammatory effects. Therefore, this study aimed to investigate the antiviral potential of T. sapiens propolis from Indonesia against SARS-CoV-2 using both in vitro and in silico methods. In silico analysis was conducted by molecular docking to evaluate the binding interactions of 30 bioactive compounds from T. sapiens propolis against RNA-dependent RNA polymerase (RdRp), a key enzyme in viral replication. The phytochemical substances found were 1,5-Dimethyl-4-[[(2-methyl-6 phenylthieno[2,3-d] pyrimidin-4yl) hydrazinylidene] methyl]pyrrole-2carbonitrile, Isocalopolyanic acid, Glyasperin A, Yucalexin B7, and Sulabiroin A. Additionally, in vitro experiments assessed the efficacy of T. sapiens propolis in inhibiting viral activity in Vero E6 cells. The results showed that propolis extract had virucidal properties, directly inactivating the virus. This study identified T. sapiens propolis as a promising natural source for antiviral drug development and future therapeutic applications.
Antiviral; In silico; In vitro; Propolis; RdRp; SARS-CoV-2; Vero E6 cells
The outbreaks of syndrome coronavirus (SARS-CoV-2), the virus responsible for COVID-19, are associated with global health crisis underscoring the need for innovative antiviral therapies (Hoenigl et al., 2022). Although COVID-19 is no longer classified as a pandemic, the virus remains a significant public health concern, particularly in vulnerable populations and regions with limited access to healthcare(Alifia et al., 2022; Borovkov et al., 2022). The continued prevalence of SARS-CoV-2 shows the importance of identifying broad-spectrum antiviral agents that can inhibit viral replication, reduce disease severity, and mitigate future outbreaks of related pathogens (Robinson et al., 2022; Tunjung et al., 2020).
In this context, natural products such as propolis, have gained attention for their potential as antiviral agents due to rich composition of bioactive compounds with diverse pharmacological activities (Berretta et al., 2020; Kumar et al., 2020; Scorza et al., 2020). This type of bee product has shown activity against viral replication and immune modulation (Pratami et al., 2020a; 2020b). Previous studies have provided evidence of the antiviral activity of propolis from Apis mellifera and Tetragonula laeviceps against viruses such as herpes simplex and influenza. Propolis from A. mellifera has been applied in formulations to treat SARS-Cov2 infections (Berretta et al., 2020).
Tetragonula sapiens is a stingless bee within the genus Tetragonula, which comprises approximately 31 species found across Oceania, including countries such as Australia, Indonesia, and Malaysia. Propolis from T. sapiens is known for its rich composition of bioactive compounds, including flavonoids, phenolic acids, and terpenoids (Asih et al., 2022a). Propolis is a natural resin resembling wax, generated by honeybees (A. mellifera L.), comprising salivary secretions, pollen, and several plant materials. Honeybees use propolis as a form of adhesive, often referred to as bee glue, to seal gaps and openings within their hives (Pavlovic et al., 2020). This practice deters parasitic intrusions and aids in regulating the internal temperature as well as humidity of the hive. The compounds produced contribute to its remarkable pharmacological properties, such as antimicrobial, antioxidant, and anti-inflammatory effects (Asih et al., 2022a; Sahlan et al., 2021; 2019). Specifically, T. sapiens propolis has shown potential antiviral activity, attributed to the ability to inhibit viral replication and modulate immune responses. Previous studies reported the efficacy of T. sapiens propolis against a range of viruses by targeting critical viral proteins and disrupting replication mechanisms. Propolis and its various components have shown significant preclinical effectiveness as antiviral agents including adenoviruses, influenza, respiratory tract, herpes simplex virus types 1 and 2 (HSV-1 and HSV-2), human immunodeficiency virus (HIV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Yosri et al., 2021). For example, molecular docking studies reported that T. sapiens propolis compounds could bind effectively to viral enzymes, including RNA-dependent RNA polymerase (RdRp), a key enzyme in viral replication (Sahlan et al., 2021), SARS-CoV-2 spike protein (Sahlan et al., 2023), and PAK-1 enzyme (Asih et al., 2022b). However, limited data exist on the specific antiviral properties of T. sapiens propolis, particularly in the context of SARS-CoV-2.
Based on the description, this study aimed to investigate the antiviral activities of T. sapiens propolis from Indonesia using in vitro and in silico methods. The investigation was carried out on the anti-SARS-CoV-2 activity in vitro of standardized herbal preparations. The results showed that propolis extract was a virucidal agent that could directly inactivate the viral particle. Furthermore, molecular docking study was carried out as a computational method that could simulate the most optimal position of the ligand-receptor complex, using the minimum scoring function or the free energy of the entire system (Husnawati et al., 2023; Mohanty and Mohanty, 2023). Since its introduction in the 1970s, the method served as reliable strategy for understanding chemical interactions and facilitating the identification of novel drugs (Pinzi and Rastelli, 2019). In a previous investigation conducted by Miyata et al. (2020; 2019), a total of 13 resident compounds of T. sapiens propolis were discovered (Miyata et al., 2020; 2019). These compounds along with an additional 17 identified through the liquid chromatography-mass spectrometry (LC-MS) method, were evaluated using molecular docking to specifically target the RdRp protein (PDB ID: 7BV2). RdRp was identified as an enzyme playing a significant role in the replication of the viral genome and the transcription of genetic material (Hillen et al., 2020). The molecular docking evaluation included analyzing the docking score, inhibition constant, and interaction profile. An in vitro experiment was also conducted to focus on evaluating the efficacy of T. sapiens propolis against SARS-CoV-2 in Vero E6 cells. By combining the methods, this study provided insights into the potential mechanisms of action and showed the significance of T. sapiens propolis as a natural source for antiviral drug development. The results were also expected to serve as the basis for further exploration of propolis-based therapies to address current and future viral threats.
2.1. Samples
Propolis extract from T. sapiens (North Luwu district, South Sulawesi, Indonesia) was macerated by ethanol and dried with an industrial spray dryer at Phytochemindo Reksa Co (Bogor, West Java, Indonesia). Spray-dried propolis (SDP) was evaluated to fulfill standardized herbal preparations with CoA No 20221011-1S.1-1P024SDP-001. This sample was applied for in vitro evaluation using Vero E6 cells.
2.2. Cell Lines and Viruses
Vero E6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, BRL, Gaithersburg, MD, USA) supplemented with 10% (w/v) fetal bovine serum (FBS). The cell lines were acquired from the Chang Gung Memorial Hospital, with passages less than 25. The SARS-CoV-2 strain CDC-4 (CGMH-CGU-01; GISAID accession number EPI_ISL_411915 and NCBI accession number MT192759) was obtained from the Taiwan Center for Disease Control and replicated in Vero E6 cells. Subsequently, SARS-CoV-2 was managed in a biosafety level three (BSL3) facility at the Institute of Preventive Medicine, National Defense Medical Center, Taiwan.
2.3. Anti-SARS-CoV-2 In Vitro
A total of two tests were performed, namely the cytotoxicity test of propolis and propolis activity against SARS-CoV-2. For the cytotoxicity test, the method by (Tang et al., 2021) was used. Initially, a 96-well tissue culture plate was seeded with Vero E6 cells (2 × 104/well) and incubated at 37oC for 16–20 h under 5% CO2. Cell viability was measured following incubation with SDP for five days, fixed by 4%PFA, and stained with 0.5% (w/v) crystal violet for 30 min. Moreover, CC50 represented the cytotoxicity concentration at which 50% of cell death occurred.
Vero E6 cells were seeded at density 2 × 104 per well in 96-well plates. These cells were infected with SARS-CoV-2 at a multiplicity of infections (MOI) of 2.5 × 104 plaque forming unit (PFU) per cell, equivalent to the TCID 50 (median tissue culture infective dose). Subsequently, cells were cultured in E2 medium comprising DMEM supplemented with 2% (v/v) FBS, alongside varying concentrations of SDP. Cells viability was evaluated after incubating at 37°C with 5% CO2 for 72 hours. The anti-SARS-CoV-2 of SDP was managed in a BSL3 laboratory at the Institute of Preventive Medicine, National Defense Medical Center, Taiwan. The overall method of in vitro evaluation is shown in Figure 1.
Figure 1 Illustration scheme of the Anti-SARS-Cov-2 In Vitro Study
2.4. Phytochemical Compounds of T. sapiens Propolis
The initial phase of the in silico study or molecular docking included the preparation of the ligand candidate. A total of 17 compounds obtained from the LC-MS and 13 T. sapiens propolis compounds from South Sulawesi published by Miyata (Miyata et al., 2020; 2019) were used as ligands. The details of these 30 compounds are presented in Table 1 (Supplementary File). All the compounds were drawn using Marvin Sketch 20.8 software (Laxmi, 2022) and the overall method of in silico study was shown in Figure 2.
Figure 2 Illustration scheme of molecular docking process
2.5. Main Protein Preparation
The protein file comprising an RdRp complex with its natural ligand, remdesivir, was downloaded in .pdb format from https://www.rcsb.org/ using PDB ID:7BV2. This complex was separated using visual molecular dynamics, VMD software (Spivak et al., 2023; Humphrey et al., 1996) and saved in .pdb format. Furthermore, the file was edited using Notepad++ software (https://notepad-plus-plus.org/). The polar hydrogen was added to the main protein using AutoDock 1.5.6 software (Forli et al., 2016).
2.6. Ligands Evaluation Through Lipinski Rule of Five & SwissADME and Ligand Preparation
The 2D and 3D structures of test compounds in Figures 3 and 4, were created using MarvinSketch. The compounds were also saved in SMILES format for Lipinski Rules of Five (Lipinski RO5) evaluation. This analysis was performed by entering the SMILES format file into webtool at https://scfbio-iitd.res.in/(Jayaram et al., 2012; Lipinski, 2004) or SwissADME webtool at http://www.swissadme.ch/ (Daina et al., 2017) to determine their drug-like properties and bioavailability.
Figure 3 Seventeen (17) compounds from Tetragonula sapiens resulted from Liquid chromatography-mass spectrometry (LC-MS)
The Lipinski RO5 consists of four specific criteria, namely a molecular weight (MW) of less than 500 g/mol, a LogP value below five, fewer than five hydrogen bond donors, and a maximum of 10 hydrogen bond acceptors (Karami et al., 2022). For molecules to be considered adequate drug-like properties and bioavailability, two out of these four defined criteria must be fulfilled. After passing the Lipinski RO5, the preparation was performed in AutoDock 1.5.6 software. In the process of preparing protein input files, all water molecules, ligands, and ions were eliminated. This was followed by the addition of polar hydrogens from the PDB file using the prepare_receptor4.py command of the AutoDock 1.5.6 software.
Figure 4 13 compounds from Tetragonula sapiens published by (Miyata et al., 2020; 2019)
2.7. Validation of Molecular Docking Method
The AutoDock Vina program needs docking parameter in order to run. The docking parameters consist of grid box dimensions and center coordinates (Eberhardt et al., 2021). In this validation process, conformation of the natural ligand to the receptor in the experimental crystallographic structure was compared with redocked samples using AutoDock software. This would be accomplished by configuring the Grid box dimensions (x, y, z), center coordinates (x, y, z), and default spacing. The outcomes of the assessment were quantified using the root mean square deviation (RMSD) value. Moreover, the docking method was considered valid when the RMSD value was less than or equal to 2 Å (Maden et al., 2023). When the RMSD value obtained was greater than 2 Å, the procedure used was considered invalid. This showed the need for manual adjustments to the Grid box dimensions and center coordinates until an RMSD of 2 Å or less was achieved (Husnawati et al., 2023; Amrulloh et al., 2023).
Table 1 The Assessment of Drug likeness of a Compound Utilizing Lipinski Rule of Five (RO5) and Swiss-Absorption, Distribution, Metabolism, and Excretion (SwissADME) Web Application
Information: Yes = Meets the requirements of Lipinski RO5 and/or SwissADME, No = Does not meet the requirements, = complies with Lipinski’s rule, = does not comply with Lipinski's rule
In this experiment, the docking coordinates in the grid box menu were formatted in a 'centered on ligand' manner, with an A1 spacing. The grid box dimensions were given as . Additionally, the redocking operation yielded 10 docking poses together with their corresponding binding affinity values. The pose with the lowest binding affinity value was selected, and its coordinate was assessed for similarity to the empirically derived model by calculating the RMSD using PyMOLL (Schrödinger, inc., USA)(Yuan et al., 2017). Therefore, the simulated form accurately represented the docking position between RdRp and remdesivir, which was initially generated from https://www.rcsb.org/ (PDB ID: 7BV2).
2.8. Docking Process
Semi-flexible docking was performed in this study, where the receptor was used as a rigid object, and ligand as a flexible object (Li et al., 2022; Zhang et al., 2022). The AutoDock Vina software was used to conduct molecular docking. The dimensions and coordinates of the grid box were acquired from the preceding step. The genetic algorithm parameters were set to their default values, with a population size of 150, a medium maximum evaluation, a maximal number of generations of 27,000, and the top automatically survived individual set to 1. Furthermore, the precise values for gene mutation and crossover rates were indicated as 0.02 and 0.8, respectively. The remaining docking parameters were changed back to their normal settings.
In this procedure, the 30 ligand compounds including RdRp-positive control, were docked individually with the RdRp protein. The docking process generated different binding poses and scores were generated, showing the strength of affinity between a ligand and its receptor or target protein. Docking scores were expressed in binding free energy and the empirical equation model prepared by Morries et al was presented in Equation 2 (Eberhardt et al., 2021; Du et al., 2016; Quiroga and Villarreal, 2016)
The first four terms of equation 2 represent molecular mechanics such as dispersion/repulsion, hydrogen bonding, electrostatics, and deviation from covalent geometry. is modeling rotor internals and global rotation and translation. Meanwhile,
is modeling desolvation on bonds and hydrophobic effects (Eberhardt et al., 2021; Quiroga and Villarreal, 2016).
2.9. Inhibitory Constant Calculation
The derived from the molecular docking results can be modified to determine the inhibitory constant (Ki) (Srinivasan, 2023; Ortiz et al., 2019; Bearne, 2012). This coefficient is defined as the dissociation constant (Kd) of the enzyme-inhibitor complex, as opposed to the binding constant (Kb). The unit of Ki is M or
and derivation of Ki value is shown in equations 3 to 6. Where R = universal gas constant (1.987 cal/mol K), and T = absolute temperature in Kelvin, with T=298.15 K (El-Hadi et al., 2020; Hawwa et al., 2009).
2.10. Analysis and Visualization
The best accurate anticipated poses were acquired from the docking simulation and examined using PyMol (Schrödinger, Inc., New York, NY, USA) (Yuan et al., 2017). Furthermore, the LigPlot+ software (The European Bioinformatics Institute, United Kingdom) was used to create 2D schematic representations of the ligand and the residues that interacted with it in the binding site of the receptor (Laskowski and Swindells, 2011).
3.1. SDP Shows Anti-SARS-CoV-2 Activity In Vero E6 Cells
The cells viability observed in this study is shown in Figure 5. The series concentration of SDP was 0, 1.17, 2.34, 4.69, 9.38, 18.75, 37.50, and 75.00 µg/ml, with viability cells of 90-100 %. The crystal violet (CV) cells cytotoxicity assay used the color of crystal violet to evaluate cells viability or drug cytotoxicity, serving as a widely used method. Moreover, a simple method for assessing the continued adhesion of Vero E6 cells is by staining the attached cells with crystal violet dye, which has an affinity for proteins and DNA. Vero E6 cells that experience death show a loss of adhesion and are eliminated from the cell population, causing a decrease in the quantity of crystal violet staining in a culture.
Figure 5 Cytotoxicity of SDP in Vero E6 cells
The potential activity of SDP against SARS-CoV-2 is shown in Figure 6. The purple color is attributed to highly diluted virus stock, indicating a small or absence of virus present. This is common in antiviral screening assays when working with very dilute virus solutions. The Vero E6 cells were seeded at density 2 × 104 per well and the cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 2.5 × 104 PFU per cell. This showed there were five infectious viral particles per well resulting in purple color. In the top five wells of column A, the cells were completely killed by the amount of virus. However, 37.5 µg/ml propolis protected four out of five wells (column B), and 18.8 µg/ml propolis showed dose-dependent protection in two (column C).
Figure 6 The activity of SDP against SARS-Cov-2. A: Mock; B: Propolis 37.5 µg/ml; C: Propolis 18.8 µg/ml
The SDP has anti-ARS-CoV-2 activity at 37.5 µmg/ml, which protected the Vero E6 cells from SARS-CoV-2 infection compared to mock control (column A). However, the SDP 18.88 µg/ml had lower activity compared to SARS-CoV-2. As shown in column C, there were 2 wells consisting of dead adherent cells and 2 wells comprised normal Vero E6 cells. The optimum dose of SDP 75 µg/ml also showed good viability cells, not toxic for Vero E6 cells. Therefore, this concentration was recommended as a therapeutic dose for the next study (pre-clinical or clinical trial). This is the first report of potential SDP against SARS-CoV-2 in vitro study.
3.2. Evaluation of Lipinski Rule of Five & SwissADME
Before conducting molecular docking, the test compounds were initially selected based on the Lipinski RO5 (Karami et al., 2022; Chagas et al., 2018). This process was carried out to evaluate certain specific parameters, such as absorption, distribution, metabolism, and excretion, affecting the pharmacokinetic properties in the human body (Karami et al., 2022; Chagas et al., 2018; Veber et al., 2002). Lipinski RO5 comprises four criteria, namely MW<500g/mol, LogP<5, H Donor<5, and For molecules to be regarded as having sufficient drug-like and bioavailability, two out of the four established requirements must be fulfilled (Karami et al., 2022; Quimque et al., 2021).
Another webtool to evaluate drug-like is SwissADME which can be accessed at http://www.swissadme.ch/. This webtool offers a web-based option for assessing the drug-like properties and bioavailability of test substances (Bakchi et al., 2022; Daina et al., 2017). Furthermore, SwissADME enables the computation of key physicochemical, pharmacokinetic, drug-like, and related parameters for one or multiple molecules(Daina et al., 2017). The evaluation entails inputting SMILES data for each phytochemical molecule into the http://www.swissadme.ch/ website to generate absorption, distribution, metabolism, and excretion results, confirming the compliance of compounds to all the requirements as drug.
All 30 propolis compounds comprise both Lipinski RO5 and SwissADME indicating the drug-like, as shown in Table 1. The detailed value of Lipinski parameters in each compound is shown in Supplementary File Table 2. The concept of drug-like is primarily focused on oral medications, as the route of administration is among the most prevalent in clinical settings. Oral drugs offer ease of use for patients, without specialized medical intervention, and typically present a higher level of convenience compared to alternative methods (Lou et al., 2023).
3.3. Root Mean Square Deviation (RMSD) Value of RedockingDocking Parameters
Following the preparation of ligand and protein structures, the docking parameter becomes a crucial input for AutoDock program. This parameter entails identifying the coordinates of the ligand-binding site on the target protein. In cases where the binding site is unknown, blind docking may be conducted by enclosing the entire protein within the grid box. As an alternative, a smaller grid box can be positioned over a specific known or predicted ligand-binding area on the protein (Maden et al., 2023; Fan et al., 2021; Quiroga and Villarreal, 2016; Feinstein and Brylinski, 2015). In this study, Gridbox dimension of 25Åx25Åx25Å was selected because it covered the main protein area.
After determining the ligand-binding region using the Gridbox in AutoDockTools, the protein coordinates were specified in the input configuration file. To obtain the data corresponding to the specified center coordinates for Gridbox, a redocking procedure was conducted between the primary protein and its native ligand. This procedure was used to confirm the docking methodology and evaluate the precision (Hevener et al., 2009). In the context of molecular docking, redocking takes place when the ligand is reinserted into the active site of the target protein following the computation of its initial energy and positioning. When the redocked ligand closely correlated with the position of the crystallographic ligand, a strong relationship was identified (Venkatesh, 2022; Hevener et al., 2009).
In this study, RdRp was redocked with remdesivir, yielding several poses. The optimal pose, characterized by the lowest docking score between RdRp and remdesivir, was analyzed using PyMol to calculate the RMSD value presented in Table 4. Furthermore, RMSD is as distance between the corresponding atoms of the ligand's docked pose and its experimental binding conformation (Ramírez and Caballero, 2018). This metric is commonly used for evaluating the accuracy of docking calculations (Shamsian et al., 2024). A high RMSD value indicates dissimilarity, while zero value shows an identical conformation structure.
The results are anticipated to meet the standards by achieving a value of (Maden et al., 2023; Trott and Olson, 2010). An RMSD value of less than or equal to 2 Å indicates a successful docking result (Shamsian et al., 2024; Maden et al., 2023; Abdalla et al., 2022). Since the RMSD value obtained in this redocking process was 2 Å, this suggested that the procedure was valid. Therefore, the data in Table 2 could be applied as the foundation for the docking procedure between RdRp and the 30 test compounds using AutoDock Tools.
Table 2 GridBox Area, Docking Coordinates, and Root Mean Square Deviation Value.
3.4. Docking Score
In this study, 30 compounds identified from T. sapiens propolis were docked to the RdRp protein through the molecular docking method, specifically using AutodockTools. The purpose of molecular docking is to predict the binding configuration and strength of a small molecule at the active site of a designated target protein (Zhang et al., 2022). The outcome is represented by a value that indicates the free-binding energy. This shows the energy needed for the ligand to establish a connection with the receptor. The free binding energy often referred to as the docking score, is used to assess the strength of the interaction between a ligand (test substance) and its receptor (target protein), which is calculated using Equation 7 (Srinivasan, 2023; Du et al., 2016):
The symbols indicate the enthalpy and entropy, respectively. In molecular docking, the objective is to achieve a negative
value. The occurrence of this situation is initiated by the process of binding, where the enthalpy decreases due to intermolecular contacts and bond formation. However, the entropy increases because of the reduction in degrees of freedom(Bearne, 2012), leading to negative value of
. A more negative value indicates a stronger affinity between the ligand and the receptor (Srinivasan, 2023; Berenger et al., 2021; Fukunishi et al., 2018; Trott and Olson, 2010).
Table 3 presents the docking scores for 30 compounds in relation to RdRp, with remdesivir, the natural ligand of RdRp, serving as the positive control. Remdesivir, a recently FDA-approved SARS-CoV-2 drug (Chera and Tanca, 2022) has been shown to target RdRp with a low binding energy of -6.6 kcal/mol. Based on the analysis, only 10 out of the 30 test compounds achieved a docking score that was lower than remdesivir. This suggested that the 10 compounds had a greater affinity for RdRp compared to remdesivir. Specifically, the 10 were 1,5-Dimethyl-4-[[(2-methyl-6-phenylthieno[2,3-d]pyrimidin-4-yl)hydrazinylidene]methyl]pyrrole-2-carbonitrile, isocalopolyanic acid, glyasperin A, Yucalexin B7, Sulabiroin A, Oleandrigenin, broussoflavonol F, Glyurallin B, Mollicellin H, and Dulxanthone. This phenomenon showed the potential application of propolis compounds as RdRp inhibitors.
By comparing the docking scores or binding energy with other substances against RdRp, the propolis compounds showed significant affinity. A previous study conducted by Hosseini used AutoDock Vina to evaluate 12 FDA-approved drugs against RdRp. The results identified five compounds with the lowest docking scores or binding energy. These included Leucal (Leucovorin) at -8.2 kcal/mol, Natamycin -7.8 kcal/mol, Isavuconazonium -7.2 kcal/mol, and Folinic acid also -7.2 kcal/mol (Hosseini et al., 2021). Additionally, another molecular docking study using AutoDockTools 1.5.6 and AutoDock Vina assessed mangosteen compounds against RdRp. The results showed a lowest binding energy of -7.8 kcal/mol (Afladhanti et al., 2022), which was slightly lower than the PS06 compound (1,5-Dimethyl-4-[[(2-methyl-6-phenylthieno[2,3-d]pyrimidin-4-yl)hydrazinylidene]methyl]pyrrole-2-carbonitrile), with a docking score of -7.9 kcal/mol. Despite the use of the same software, the binding energy values obtained in this study cannot be directly compared with other reports due to the differing docking parameters, such as Gridbox dimensions and center coordinates. However, this analysis indicates that the compounds found in T. sapiens propolis show potential as inhibitors of RdRp.
The value from the molecular docking results can be altered to generate an inhibition constant (Ki) for ligand and receptor interaction using equation 5. This coefficient serves as a measure of potency of an inhibitor. Table 3 shows the Ki values for each pair of primary proteins and the test compounds. The inhibition constant, also known as the half-maximal inhibitory concentration, is a measure of potency. It represents the concentration needed to achieve 50% inhibition of the maximum effect (Ortiz et al., 2019). However, with an increasing negative docking score, the inhibition constant becomes minimal. This shows a stronger inhibition of the test compounds against the target protein.
Table 3 Docking Score & Ki between RdRp and the Test Compounds
1 Ki=Inhibition constant
3.5. Interaction Profiles
Ligplot+ was used to visually profile the 2D bonds and examine the interactions occurring in the docking area below 5 Å, specifically focusing on hydrogen bonds and hydrophobic processes. Figure 7 shows the interactions between RdRp and the five ligands with the lowest docking scores, as well as RdRP and its original ligand, remdesivir, correspondingly. The bonds that formed the atoms in ligand were purple, while brown was obtained in the receptor. Subsequently, the ligand and receptor were connected by a green dotted line, which symbolized a hydrogen bond. The numerical value denoted the length of this link, while hydrophobic interaction was represented by a brown fan-shaped structure. Furthermore, the ligand atoms were identified by the presence of reddish-brown luminescent lines. Another advantage of the Ligplot+ program is that the atoms are distinguished by color for easy identification (Laskowski and Swindells, 2011; Wallace et al., 1995). The colors blue, black, yellow, red, and purple corresponded to the element’s nitrogen, carbon, sulfur, oxygen, and phosphorus, respectively.
Based on Ligplot+ 2D visualization results, the RdRp complex with remdesivir obtained seven hydrogen bonds from six separate amino acids and eight hydrophobic interactions. These abundant hydrogen bonds show a strong and stable connection, thereby contributing the most negative weight to the docking score, with a high binding energy.
Several key amino acid residues, including Asp760, Thr687, Ala688, Lys545 and Val557, were observed in RdRp active site (Kumar et al., 2020). This site was where the substrate experienced a catalyzed chemical reaction. In this study, the RdRp enzyme accelerated the replication process of SARS-CoV-2. The active site also acts as an important target zone for binding with inhibitors to indirectly prevent virus replication. Furthermore, the active site between SARS-CoV-2 and SARS-CoV that instigated SARS appears identical in terms of amino acid residues but certain conformational differences occurred that contributed to binding specificity. The docking activities in this study are conducted on RdRp active sites.
Various key residues, including Ser759, Asp760, Asp761, Lys545, and Arg555 occurred in RdRp catalytic site (Kumar et al., 2020). This site refers to a specific area on an enzyme molecule where the actual reaction takes place. Catalytic residues of the site interact with the substrate to lower the activation energy of a reaction, thereby accelerating the process. Additionally, the catalytic site is where the amino acid residues intend to catalyze the substrate.
The evaluation process considered the bond or interaction distance. This interval particularly in hydrogen bonds, serves as an analytical indicator to represent the interaction strength (Trott and Olson, 2010). In general, hydrogen bonds in proteins exist in the moderate category. The bond distance is classified into three groups, depending on the intensity level (Buckingham et al., 2008). These ranges include (medium/ionic or electrostatic as well as
(weak and also electrostatic). Therefore, closer distances tend to generate stronger bonds.
Based on the results, 1,5-Dimethyl-4-[[(2-methyl-6-phenylthieno[2,3-d] pyrimidine-4-yl) hydrazinylidene] methyl] pyrrole-2-carbonitrile (PS06) showed similar interaction profiles. Consequently, RdRp interaction profiles of approximately 86.7% showed sufficient similarity. A total of 13 out of the 15 residues appeared similar to the counterparts in the positive control, namely the remdesivir inhibitor against RdRp. The PS06 compound only formed a single hydrogen bond with Thr680 at a distance of indicating a weak category. This suggested that out of 14 amino acid residues that reacted hydrophobically with PS06, only Thr687 and Ala688, interacted from RdRp active site, while Arg555 was observed from the catalytic site. The results show that several interactions with important amino acid residues are possible, with the maximum percentage similarity of the interaction profile against the positive control at 86.7%. Moreover, this study recommends further development of the compound (PS06) as a promising RdRp inhibitor.
Isocalopolyanic acid (PK07) did not show any similarity in the interaction profile with the RdRp complex and remdesivir. The acid compound formed two hydrogen bonds with Asn497 and Arg569 residues at a distance of respectively, indicating a moderate bond strength. Subsequently, out of the eight amino acid residues that reacted hydrophobically with the isocalopolyanic acids, only Val557 interacted from RdRp active site. This result is due to the lack of similarity in the interaction profile with the positive control. Therefore, isocalopolyanic acid compounds are not recommended for further development as RdRp inhibitors.
Glyasperin A (PK08) showed a similar interaction profile with RdRp complex and remdesivir. However, only one out of 11 interacting amino acid residues was identical to the positive control. This PK08 compound formed one single and two hydrogen bonds with Asp760 (key residue) and Ser814 at a distance of 2.82, 2.87, and respectively, indicating a moderate bond strength. The results showed that glyasperin A was strongly bonded to RdRp, due to the influence on bond stability. Glyasperin A obtained the most negative weight in the scoring function using AutodockVina software. This condition generated significant binding energy. Subsequently, out of the 8 amino acid residues that reacted hydrophobically with glyasperin A, only Asp761 and Ser75 interacted from the RdRp catalytic site. Despite showing a minimal percentage similarity of 6.7%, glyasperin A compounds are possibly considered for further studies as a potential RdRp inhibitor, due to the interactions with key residues.
Figure 7 Two-Dimensional (2D) Interaction Profile of RdRp against: (a) 1,5-Dimethyl-4-[[(2-methyl-6 phenylthieno[ 2,3-d] pyrimidin-4 yl) hydrazinylidene] methyl]pyrrole-2 carbonitrile; (b) Isocalopolyanic acid; (c) Glyasperin A; (d) Yucalexin B7; (e) Sulabiroin A; (f) Remdesivir
Yucalexin B7 (PS04) did not show any similarity in the interaction profile with the RdRp complex and remdesivir. This compound formed a single hydrogen bond with Arg569 at a distance of 3.12, indicating a moderate bond strength. Among the seven amino acid residues that reacted hydrophobically with the Yucalexin B7, only Val557 interacted from the RdRp active site. Therefore, due to the lack of similarity in the interaction profile with the positive control, isocalopolyanic acid compounds were not recommended for further development as RdRp inhibitors.
Sulabiroin A (PK01) did not show any similar interaction profile with the RdRp complex and remdesivir. This compound formed three hydrogen bonds with Lys545 (key residue), Ser501, and Val560 at 3.01, 3.23 and respectively, indicating a moderate bond strength. Based on the interaction with the key residue, a strong bond between the sulabiroin compound A and the RdRp active site was found. Among the eight amino acid residues that combined hydrophobically with Sulabiroin A, only Val557 interacted from RdRp active site. The presence of two interactions with amino acids also showed significant potential for sulabiroin A compounds as RdRp inhibitors. Consequently, sulabiroin A could be considered for advanced studies due to the 0% similarity of the interaction profile in the compounds.
Based on the results, 1,5-Dimethyl-4-[[(2-methyl-6-phenylthieno[2,3-d] pyrimidin4yl) hydrazinylidene] methyl] pyrrole-2-carbonitrile (PK06) showed the most potential as RdRp inhibitors among the 30 propolis compounds. These results were obtained after the assessments of docking score inhibition constant (Ki), and interaction profile. Figure 8 represents the 3D visualization results using PyMol from PK06 and a positive control inhibitor (remdesivir) in a similar binding pocket of RdRp.
Further analysis was conducted to determine the relevance of the information obtained in terms of developing COVID-19 therapeutics. This included applying five propolis compounds with the most potential after purification or the complete samples because the information was used as a claim. However, in providing solutions to these questions, a literature study should be conducted. The objective is to analyze the comparison of including the entire propolis to the samples showing certain biological characteristics. Therefore, there is a need to validate the occurrence of possible synergistic effects on the propolis compounds.
Figure 8 Three-dimensional (3D) visualization using the PyMol from PK06 (red) and remdesivir (yellow) in the similar binding pocket in the RdRp enzyme
Synergy refers to the significant effect of combining several compounds, compared to the contribution of individual components. This condition implies that in synergistic cases, additional effectiveness is obtained by using a combination of compounds rather than independent application. A previous study evaluated the impact on antioxidant activity with five main flavonoid compounds in propolis, compared to the complete samples (Osés et al., 2020). The results showed that the use of the entire mass was able to suppress the formation of oxidants or free radicals, compared to individual or combined applications. Another study showed that using propolis in its entirety had significant antiviral activity against HSV-1 than flavone compounds separately or in combination (Amoros et al., 1992).
Several studies have focused on the synergistic effects of many propolis components, as opposed to their single application. For example, the use of three flavonoid compounds derived from propolis caused greater antimicrobial efficacy compared to the individual components (Kharsany et al., 2019). Additionally, investigations examined the synergism of propolis in combination with other components. The combined impact of propolis-honey has also been evaluated on the antimicrobial activity against C. albicans, S. aureus, and E. coli. The results showed that the combined sample generated extensive antimicrobial characteristics, compared to the individual elements (Noori et al., 2012). The combination of propolis and bee venom demonstrated higher anti-proliferative behavior on breast cancer cells than with separate usage (Drigla et al., 2016). Regardless of previous studies that obtained the five most potential propolis compounds for COVID-19 treatment, propolis is currently applied as a therapeutic agent in its entirety rather than for individual application. However, for validation purposes, further studies should be conducted, including molecular dynamic analysis and in vivo testing appear necessary.
In conclusion, the evaluation results of in vitro study showed that SDP at 37.5 µg/ml had anti-SARS-CoV-2 activity using Vero E6 cell-infected viruses. The molecular docking obtained the five best propolis compounds from each target protein. The results showed that based on docking score and inhibition constants, T. sapiens propolis could block the RdRp enzyme, thereby reducing viral loads. According to literature studies, the samples showed substantial synergistic effects that were collectively applied to generate extensive biological activities, compared to individual engagement. Furthermore, the observation confirmed that the potential for propolis compounds in developing antiviral COVID-19 therapeutics greatly depended on the absolute utilization of the samples.
The authors are grateful for the several financial supports partly provided by the Rispro United Kingdom-Indonesia Consortium on Interdisciplinary Sciences (UKICIS) (Contract Number: 4345/E4/AL.04/2022) from LPDP and Ministry of Education, Culture, Research and Technology of The Republic of Indonesia; Faculty member seed fund (Contract number: NKB-3438/UN2.F4.D/PPM.00.00/2024) from Faculty of Engineering, University of Indonesia; the National Science and Technology Council, Taiwan (NSTC 113-2321-B-182-003, 113-2320-B-182-012-, and 112-2320-B-182-034-MY3); Chang Gung Memorial Hospital (BMRP416 and CMRPD1M0881-3); and the Research Center for Emerging Viral Infections from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan. Furthermore, the authors are grateful for the experimental method and BSL-3 laboratory support provided by the Institute of Preventive Medicine, National Defense Medical Center.
Author Contributions
S.C.A., M.S., and M.S. designed and conceptualized the molecular docking project. J.T.H designed and conceptualized the in-vitro experiment. S.C.A. wrote the manuscript. S.C.A and D.K.P edited the manuscript. S.C.A conducted the molecular docking experiment and analysis. J.T.H, H.P.T, S.K.T, and C.H.C conducted the In-vitro experiment and analysis. M.S., M.N., and A.M. supervised. M.S. and J.T.H managed the funding. All authors have given approval to the final version of the manuscript.
Conflict of Interest
The authors have no conflicts of interest to declare. All co-authors have seen and agreed with the contents of the manuscript and there is no financial interest to report.
| Filename | Description |
|---|---|
| R2-CE-7192-20250219113753.docx | --- |
| R2-CE-7192-20250314145718.docx | --- |
Abdalla, M, Eltayb, WA, El-Arabey, AA, Singh, K & Jiang, X 2022, 'Molecular dynamic study of SARS-CoV-2 with various S protein mutations and their effect on thermodynamic properties', Computers in Biology and Medicine, vol. 141, article 105025, https://doi.org/10.1016/j.compbiomed.2021.105025
Afladhanti, PM, Romadhan, MD, Hamzah, HA & Bhelqis, Q, 2022, ‘Molecular Docking Study of Garcinia mangostana (Mangosteen) Compounds as SARS-CoV-2 Potential Inhibitors’ Sriwijaya Journal of Medicine, vol. 5, no. 1, pp. 31–40, https://doi.org/10.32539/SJM.v5i1.127
Alifia, KCH, Kontoravdi, C, Kis, Z & Ismail, D 2022, 'Techno-economic evaluation of novel SARS-CoV-2 vaccine manufacturing in the insect cell baculovirus platform', International Journal of Technology, vol. 13, no. 8, pp. 1630-1639, https://doi.org/10.14716/ijtech.v13i8.6139
Amoros, M, Simõs, CMO, Girre, L, Sauvager, F & Cormier, M 1992, 'Synergistic effect of flavones and flavonols against herpes simplex virus type 1 in cell culture. Comparison with the antiviral activity of propolis', Journal of Natural Products, vol. 55, no. 12, pp. 1732–1740
Amrulloh, L, Harmastuti, N, Prasetiyo, A & Herowati, R 2023, 'Analysis of molecular docking and dynamics simulation of mahogany (Swietenia macrophylla king) compounds against the PLpro Enzyme SARS-COV-2', Jurnal Farmasi dan Ilmu Kefarmasian Indonesia, vol. 10, no. 3, pp. 347–359, https://doi.org/10.20473/jfiki.v10i32023.347-359
Asih, SC, Pratami, DK, Yohda, M, Fukutami, Y, Faried, A & Sahlan, M 2022a, 'The Role of Propolis Tetragonula sp. in Oxidative Stress and Its Protective Effect Against UV Radiation on Cells', International Journal of Applied Pharmaceutics, pp. 123–128, https://doi.org/10.22159/ijap.2022.v14s3.26
Asih, SC, Sahlan, M & Nasikin, M 2022b, 'Preliminary study for COVID-19 drug discovery of 30 phytochemical compounds from Tetragonula sp. Propolis as PAK1 inhibitor', International Journal of Applied Pharmaceutics, pp. 116–122, https://doi.org/10.22159/ijap.2022.v14s3.25
Bakchi, B, Krishna, AD, Sreecharan, E, Ganesh, VBJ, Niharika, M, Maharshi, S, Puttagunta, SB, Sigalapalli, DK, Bhandare, RR & Shaik, AB 2022, 'An overview on applications of SwissADME web tool in the design and development of anticancer, antitubercular and antimicrobial agents: A medicinal chemist’s perspective', Journal of Molecular Structure, vol. 1259, article 132712, https://doi.org/10.1016/j.molstruc.2022.132712
Bearne, SL 2012, 'Illustrating enzyme inhibition using gibbs energy profiles', Journal of Chemical Education, vol. 89, no. 6, pp. 732–737, https://doi.org/10.1021/ed200395n
Berenger, F, Kumar, A, Zhang, KYJ & Yamanishi, Y 2021, 'Lean-docking: exploiting ligands’ predicted docking scores to accelerate molecular docking', Journal of Chemical Information and Modeling, vol. 61, no. 5, pp. 2341–2352, https://doi.org/10.1021/acs.jcim.0c01452
Berretta, AA, Silveira, MAD, Capcha, JMC & De Jong, D 2020, 'Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease', Biomedicine & Pharmacotherapy, vol. 131, article 110622
Borovkov, AI, Bolsunovskaya, MV, Gintciak, AM, Rakova, VV, Efremova, MO & Akbarov, RB 2022, 'COVID-19 spread modeling considering vaccination and re-morbidity', International Journal of Technology, vol. 13, no. 7, pp. 1463-1472, https://doi.org/10.14716/ijtech.v13i7.6186
Buckingham, AD, Del Bene, JE & McDowell, SAC 2008, 'The hydrogen bond', Chemical Physics Letters, vol. 463, no. 1–3, pp. 1–10
Chagas, CM, Moss, S & Alisaraie, L 2018, 'Drug metabolites and their effects on the development of adverse reactions: revisiting Lipinski’s rule of five', International Journal of Pharmaceutics, vol. 549, no. 1–2, pp. 133–149, https://doi.org/10.1016/j.ijpharm.2018.07.046
Chera, A & Tanca, A 2022, 'Remdesivir: the first FDA-approved anti-COVID-19 treatment for young children', Discoveries (Craiova, Romania), vol. 10, no. 2, article e151, https://doi.org/10.15190/d.2022.10
Daina, A, Michielin, O & Zoete, V 2017, 'SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules', Scientific Reports, vol. 7, no. 1, article 42717, https://doi.org/10.1038/srep42717
Drigla, F, Balacescu, O, Visan, S, Bisboaca, SE, Berindan-Neagoe, I & Marghitas, LA 2016, 'Synergistic effects induced by combined treatments of aqueous extract of propolis and venom', Clujul Medical, vol. 89, no. 1, article 104, https://doi.org/10.15386/cjmed-527
Du, X, Li, Y, Xia, YL, Ai, SM, Liang, J, Sang, P, Ji, XL & Liu, SQ 2016, 'Insights into protein–ligand interactions: mechanisms, models, and methods', International Journal of Molecular Sciences, vol. 17, no. 2, article 144, https://doi.org/10.3390/ijms17020144
Eberhardt, J, Santos-Martins, D, Tillack, AF & Forli, S 2021, 'AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings', Journal of Chemical Information and Modeling, vol. 61, no. 8, pp. 3891–3898, https://doi.org/10.1021/acs.jcim.1c00203
El-Hadi, AA, Ahmed, HM, Zaki, RA & Mohsen, AM 2020, 'Enhanced Enzymatic Activity of Streptomyces Griseoplanus L-Asparginase via its incorporation in an oil-based nanocarrier', International Journal of Applied Pharmaceutics, vol. 12, no. 5, pp. 203–210, https://doi.org/10.22159/ijap.2020v12i5.38360
Fan, M, Wang, J, Jiang, H, Feng, Y, Mahdavi, M, Madduri, K, Kandemir, MT & Dokholyan, NV 2021, 'GPU-accelerated flexible molecular docking', The Journal of Physical Chemistry B, vol. 125, no. 4, pp. 1049–1060, https://doi.org/10.1021/acs.jpcb.0c09051
Feinstein, WP & Brylinski, M 2015, 'Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets', Journal of Cheminformatics, vol. 7, p. 18, https://doi.org/10.1186/s13321-015-0067-5
Forli, S, Huey, R, Pique, ME, Sanner, MF, Goodsell, DS & Olson, AJ 2016, 'Computational protein–ligand docking and virtual drug screening with the AutoDock suite', Nature Protocols, vol. 11, no. 5, pp. 905–919, https://doi.org/10.1038/nprot.2016.051
Fukunishi, Y, Yamashita, Y, Mashimo, T & Nakamura, H 2018, 'Prediction of protein-compound binding energies from known activity data: docking-score-based method and its applications', Molecular Informatics, vol. 37, no. 6–7, article e1700120, https://doi.org/10.1002/minf.201700120
Hawwa, R, Aikens, J, Turner, RJ, Santarsiero, BD & Mesecar, AD 2009, 'Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus', Archives of Biochemistry and Biophysics, vol. 488, no. 2, pp. 109–120, https://doi.org/10.1016/j.abb.2009.06.005
Hevener, KE, Zhao, W, Ball, DM, Babaoglu, K, Qi, J, White, SW & Lee, RE 2009, 'Validation of molecular docking programs for virtual screening against dihydropteroate synthase', Journal of Chemical Information and Modeling, vol. 49, no. 2, pp. 444–460, https://doi.org/10.1021/ci800293n
Hillen, HS, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D & Cramer, P 2020, 'Structure of replicating SARS-CoV-2 polymerase', Nature, vol. 584, no. 7819, pp. 154–156, https://doi.org/10.1038/s41586-020-2368-8
Hoenigl, M, Seidel, D, Carvalho, A, Rudramurthy, SM, Arastehfar, A, Gangneux, JP, Nasir, N, Bonifaz, A, Araiza, J & Klimko, N 2022, 'The emergence of COVID-19 associated mucormycosis: a review of cases from 18 countries', The Lancet Microbe, vol. 3, no. 7, pp. 543–552
Hosseini, M, Chen, W, Xiao, D, & Wang, C 2021, ‘Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs’ Precision Clinical Medicine, vol. 4, no. 1, pp. 1–16. https://doi.org/10.1093/pcmedi/pbab001
Humphrey, W, Dalke, A & Schulten, K 1996, 'VMD: Visual molecular dynamics', Journal of Molecular Graphics, vol. 14, no. 1, pp. 33–38, https://doi.org/10.1016/0263-7855(96)00018-5
Husnawati, H, Kusmardi, K, Kurniasih, R, Hasan, AZ, Andrianto, D, Julistiono, H, Priosoeryanto, BP, Artika, IM & Salleh, MN 2023, 'Investigation of chemical compounds from phomopsis extract as anti-breast cancer using LC-MS/MS analysis, molecular docking, and molecular dynamic simulations', International Journal of Technology, vol. 14, no. 7, pp. 1476-1486, https://doi.org/10.14716/ijtech.v14i7.6696
Jayaram, B, Singh, T, Mukherjee, G, Mathur, A, Shekhar, S & Shekhar, V 2012, 'Sanjeevini: a freely accessible web-server for target directed lead molecule discovery', BMC Bioinformatics, vol. 13, S17, article S7, https://doi.org/10.1186/1471-2105-13-S17-S7
Karami, TK, Hailu, S, Feng, S, Graham, R & Gukasyan, HJ 2022, 'Eyes on Lipinski’s Rule of five: a new “rule of thumb” for physicochemical design space of ophthalmic drugs', Journal of Ocular Pharmacology and Therapeutics, vol. 38, no. 1, pp. 43–55, https://doi.org/10.1089/jop.2021.0069
Kharsany, K, Viljoen, A, Leonard, C & Van Vuuren, S 2019, 'The new buzz: investigating the antimicrobial interactions between bioactive compounds found in south african propolis', Journal of Ethnopharmacology, vol. 238, article 111867
Kumar, D, Chauhan, G, Kalra, S, Kumar, B & Gill, MS 2020, 'A perspective on potential target proteins of COVID-19: Comparison with SARS-CoV for designing new small molecules', Bioorganic Chemistry, vol. 104, p. 104326
Laskowski, RA & Swindells, MB 2011, 'LigPlot+: multiple ligand–protein interaction diagrams for drug discovery', Journal of Chemical Information and Modeling, vol. 51, no. 10, pp. 2778-2786, https://doi.org/10.1021/ci200227u
Laxmi, K 2022, 'Characterization of Ligand N’-[(1E)-1-phenylethylidene]-1, 3-benzothiazole-2-carbohydrazide by using Marvin Sketch 20.8 Software', Oriental Journal Of Chemistry, vol. 38, no. 1, pp. 77–84, https://doi.org/10.13005/ojc/380109
Li, J, Liu, G, Zhen, Z, Shen, Z, Li, S & Li, H 2022, 'Molecular docking for ligand-receptor binding process based on heterogeneous computing', Scientific Programming, vol. 2022, pp. 1–13, https://doi.org/10.1155/2022/9197606
Lipinski, CA 2004, 'Lead- and drug-like compounds: the rule-of-five revolution', Drug Discovery Today: Technologies, vol. 1, no. 4, pp. 337–341, https://doi.org/10.1016/j.ddtec.2004.11.007
Lou, J, Duan, H, Qin, Q, Teng, Z, Gan, F, Zhou, X & Zhou, X 2023, 'Advances in oral drug delivery systems: challenges and opportunities', Pharmaceutics, vol. 15, no. 2, article 484 https://doi.org/10.3390/pharmaceutics15020484
Maden, SF, Sezer, S & Acuner, SE 2023, 'Fundamentals of molecular docking and comparative analysis of protein–small-molecule docking approaches', In: Molecular Docking - Recent Advances, https://doi.org/10.5772/intechopen.105815
Miyata, R, Sahlan, M, Ishikawa, Y, Hashimoto, H, Honda, S & Kumazawa, S 2019, 'Propolis components from stingless bees collected on South Sulawesi, Indonesia, and their xanthine oxidase inhibitory activity', Journal of Natural Products, vol. 82, no. 2, pp. 205–210, https://doi.org/10.1021/acs.jnatprod.8b00541
Miyata, R, Sahlan, M, Ishikawa, Y, Hashimoto, H, Honda, S & Kumazawa, S 2020, 'propolis components and biological activities from stingless bees collected on South Sulawesi, Indonesia', HAYATI Journal of Biosciences, vol. 27, no. 1, article 82, https://doi.org/10.4308/hjb.27.1.82
Mohanty, M & Mohanty, PS 2023, 'Molecular docking in organic, inorganic, and hybrid systems: a tutorial review', Monatshefte Für Chemie - Chemical Monthly, vol. 154, no. 7, pp. 683–707, https://doi.org/10.1007/s00706-023-03076-1
Noori, AL, Al-Ghamdi, A, Ansari, MJ, Al-Attal, Y & Salom, K 2012, 'Synergistic effects of honey and propolis toward drug multi-resistant Staphylococcus aureus, Escherichia coli and Candida albicans isolates in single and polymicrobial cultures', International Journal of Medical Sciences, vol. 9, no. 9, article 793
Ortiz, CLD, Completo, GC, Nacario, RC & Nellas, RB 2019, 'Potential Inhibitors of Galactofuranosyltransferase 2 (GlfT2): Molecular docking, 3D-QSAR, and In Silico ADMETox Studies', Scientific Reports, vol. 9, no. 1, article 17096, https://doi.org/10.1038/s41598-019-52764-8
Osés, SM, Marcos, P, Azofra, P, de Pablo, A, Fernández-Muíño, MÁ & Sancho, MT 2020, 'Phenolic profile, antioxidant capacities and enzymatic inhibitory activities of propolis from different geographical areas: Needs for analytical harmonization', Antioxidants, vol. 9, no. 1, pp. 1–16, https://doi.org/10.3390/antiox9010075
Pavlovic, R, Borgonovo, G, Leoni, V, Giupponi, L, Ceciliani, G, Sala, S, Bassoli, A & Giorgi, A 2020, 'Effectiveness of Different Analytical Methods for the Characterization of Propolis: A Case of Study in Northern Italy', Molecules, vol. 25, no. 3, article 504, https://doi.org/10.3390/molecules25030504
Pinzi, L & Rastelli, G 2019, 'Molecular docking: Shifting paradigms in drug discovery', International Journal of Molecular Sciences, vol. 20, no. 18, article 4331
Pratami, DK, Indrawati, T, Istikomah, I, Farida, S & Pujianto, P 2020a, 'Antifungal activity of microcapsule propolis from Tetragonula spp. to Candida albicans', Communications in Science and Technology, vol. 5, no. 1, pp. 16–21, https://doi.org/10.21924/cst.5.1.2020.178
Pratami, DK, Mun’im, A, Hermansyah, H, Gozan, M & Sahlan, M 2020b, 'Microencapsulation optimization of propolis ethanolic extract from Tetragonula spp using response surface methodology', International Journal of Applied Pharmaceutics, vol. 12, no. 1, pp. 197–206, https://doi.org/10.22159/ijap.2020v12i4.37808
Quimque, MTJ, Notarte, KIR, Fernandez, RAT, Mendoza, MAO, Liman, RAD, Lim, JAK, Pilapil, LAE, Ong, JKH, Pastrana, AM & Khan, A 2021, 'Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms', Journal of Biomolecular Structure and Dynamics, vol. 39, no. 12, pp. 4316–4333
Quiroga, R & Villarreal, MA 2016, 'Vinardo: a scoring function based on autodock vina improves scoring, docking, and virtual screening', PloS One, vol. 11, no. 5, article e0155183, https://doi.org/10.1371/journal.pone.0155183
Ramírez, D & Caballero, J 2018, 'Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data?', Molecules, vol. 23, no. 5, article 1038, https://doi.org/10.3390/molecules23051038
Robinson, PC, Liew, DFL, Tanner, HL, Grainger, JR, Dwek, RA, Reisler, RB, Steinman, L, Feldmann, M, Ho, LP & Hussell, T 2022, 'COVID-19 therapeutics: Challenges and directions for the future', Proceedings of the National Academy of Sciences, vol. 119, no. 15, article e2119893119
Sahlan, M, Devina, A, Pratami, DK, Situmorang, H, Farida, S, Munim, A, Kusumoputro, B, Yohda, M, Faried, A & Gozan, M 2019, 'Anti-inflammatory activity of Tetragronula species from Indonesia', Saudi Journal of Biological Sciences, vol. 26, no. 7, pp. 1531–1538, https://doi.org/10.1016/j.sjbs.2018.12.008
Sahlan, M, Dewi, LK, Pratami, DK, Lischer, K & Hermansyah, H 2023, 'In silico identification of propolis compounds potential as COVID-19 drug candidates against SARS-CoV-2 spike protein', International Journal of Technology, vol. 14, no. 2, p. 387, https://doi.org/10.14716/ijtech.v14i2.5052
Sahlan, M, Irdiani, R, Flamandita, D, Aditama, R, Alfarraj, S, Ansari, MJ, Khayrani, AC, Pratami, DK & Lischer, K 2021, 'Molecular interaction analysis of Sulawesi propolis compounds with SARS-CoV-2 main protease as preliminary study for COVID-19 drug discovery', Journal of King Saud University-Science, vol. 33, no. 1, p. 101234, https://doi.org/10.1016/j.jksus.2020.101234
Scorza, CA, Gonçalves, VC, Scorza, FA, Fiorini, AC, de Almeida, ACG, Fonseca, MCM & Finsterer, J 2020, 'Propolis and coronavirus disease 2019 (COVID-19): Lessons from nature', Complementary Therapies in Clinical Practice, vol. 41, article 101227
Shamsian, S, Sokouti, B & Dastmalchi, S 2024, 'Benchmarking different docking protocols for predicting the binding poses of ligands complexed with cyclooxygenase enzymes and screening chemical libraries', BioImpacts: BI, vol. 14, no. 2, p. 29955, https://doi.org/10.34172/bi.2023.29955
Spivak, M, Stone, JE, Ribeiro, J, Saam, J, Freddolino, L, Bernardi, RC & Tajkhorshid, E 2023, 'VMD as a platform for interactive small molecule preparation and visualization in quantum and classical simulations', Journal of Chemical Information and Modeling, vol. 63, no. 15, pp. 4664–4678, https://doi.org/10.1021/acs.jcim.3c00658
Srinivasan, B 2023, 'A guide to enzyme kinetics in early drug discovery', The FEBS Journal, vol. 290, no. 9, pp. 2292–2305, https://doi.org/10.1111/febs.16404
Tang, WF, Tsai, HP, Chang, YH, Chang, TY, Hsieh, CF, Lin, CY, Lin, GH, Chen, YL, Jheng, JR, Liu, PC, Yang, CM, Chin, YF, Chen, CC, Kau, JH, Hung, YJ, Hsieh, PS & Horng, JT 2021, 'Perilla (Perilla frutescens) leaf extract inhibits SARS-CoV-2 via direct virus inactivation', Biomedical Journal, vol. 44, no. 3, pp. 293–303, https://doi.org/10.1016/j.bj.2021.01.005
Trott, O & Olson, AJ 2010, 'AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading', Journal of Computational Chemistry, vol. 31, no. 2, pp. 455–461
Tunjung, N, Kreshanti, P, Saharman, YR, Whulanza, Y, Supriadi, S, Chalid, M, Anggraeni, MI, Hamid, ARAH & Sukasah, CL 2020, 'Clinical evaluation of locally made flocked swabs in response to the COVID-19 pandemic in a developing country', International Journal of Technology, vol. 11, no. 5, article 878, https://doi.org/10.14716/ijtech.v11i5.4333
Veber, DF, Johnson, SR, Cheng, HY, Smith, BR, Ward, KW & Kopple, KD 2002, 'Molecular properties that influence the oral bioavailability of drug candidates', Journal of Medicinal Chemistry, vol. 45, no. 12, pp. 2615–2623
Venkatesh 2022, 'Molecular Docking of Ganomestenol with SARS-CoV-2 Mpro', Asian Journal of Pharmaceutical and Clinical Research, pp. 46–47, https://doi.org/10.22159/ajpcr.2022.v15i2.43679
Wallace, AC, Laskowski, RA & Thornton, JM 1995, 'LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions', Protein Engineering, Design and Selection, vol. 8, no. 2, pp. 127–134, https://doi.org/10.1093/protein/8.2.127
Yosri, N, Abd El-Wahed, AA, Ghonaim, R, Khattab, OM, Sabry, A, Ibrahim, MAA, Moustafa, MF, Guo, Z, Zou, X, Algethami, AFM, Masry, SHD, AlAjmi, MF, Afifi, HS, Khalifa, SAM & El-Seedi, HR 2021, 'Anti-Viral and Immunomodulatory Properties of Propolis: Chemical Diversity, Pharmacological Properties, Preclinical and Clinical Applications, and In Silico Potential against SARS-CoV-2', Foods (Basel, Switzerland), vol. 10, no. 8, https://doi.org/10.3390/foods10081776
Yuan, S, Chan, HCS & Hu, Z 2017, 'Using PyMOL as a platform for computational drug design', Wiley Interdisciplinary Reviews: Computational Molecular Science, vol. 7, no. 2, article e1298
Zhang, B, Li, H, Yu, K & Jin, Z 2022, 'Molecular docking-based computational platform for high-throughput virtual screening', CCF Transactions on High Performance Computing, vol. 4, no. 1, pp. 63–74, https://doi.org/10.1007/s42514-021-00086-5
