Investigation of Chemical Compounds from Phomopsis Extract as Anti-Breast Cancer Using LC-MS/MS Analysis, Molecular Docking, and Molecular Dynamic Simulations
Corresponding email: kusmardi.ms@ui.ac.id
Published at : 07 Dec 2023
Volume : IJtech
Vol 14, No 7 (2023)
DOI : https://doi.org/10.14716/ijtech.v14i7.6696
Husnawati, Kusmardi, K., Kurniasih, R., Hasan, A.E.Z., Andrianto, D., Julistiono, H., Priosoeryanto, B.P., Artika, I.M., Salleh, M.N., 2023. Investigation of Chemical Compounds from Phomopsis Extract as Anti-Breast Cancer Using LC-MS/MS Analysis, Molecular Docking, and Molecular Dynamic Simulations. International Journal of Technology. Volume 14(7), pp. 1476-1486
| Husnawati | 1. Doctoral Program of Biomedical Science, Faculty of Medicine, Universitas Indonesia, 10430, Jakarta, Indonesia, 2. Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB Unive |
| Kusmardi Kusmardi | 1. Department Anatomical Pathology, Faculty of Medicine, Universitas Indonesia, 10430, Jakarta, Indonesia. 2. Drug Development Research Center, Indonesia Medical Education and Research Institute (IMER |
| Rini Kurniasih | Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB University, 16680, Bogor, Indonesia |
| AE Zainal Hasan | Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB University, 16680, Bogor, Indonesia |
| Dimas Andrianto | Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB University, 16680, Bogor, Indonesia |
| Heddy Julistiono | Research Center for Applied Microbiology, National Research and Innovation Agency - BRIN, 16911, Bogor, Indonesia |
| Bambang Pontjo Priosoeryanto | Division of Veterinary Pathology, School of Veterinary Medicine and Biomedical Sciences, IPB University, 16680, Bogor, Indonesia |
| I Made Artika | Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB University, 16680, Bogor, Indonesia |
| Mohd Nazil Salleh | Departnent of Biomedical Sciences, Faculty of Health Sciences, University College of MAIWP International, 68100, Kuala Lumpur, Malaysia |
Since 2014, we have successfully isolated
endophytic fungi from the leaves of Indonesian Annona muricata, exhibiting
potential anti-breast cancer properties. The analysis of Internal Transcribed
Spacer (ITS) showed the identified fungi species as Phomopsis sp. The
ethyl acetate extract derived from Phomopsis sp.
inhibited MCF7 cells (IC50 <20 ppm) and reduced the number and
volume of nodules in Sprague-Dawley rats with breast
cancer. However, molecular mechanism underlying the action of
this extract in breast cancer treatment remains unclear.
Therefore, this study aimed
to identify the active compounds in Phomopsis extract
and to predict anti-breast cancer mechanism through HER2 inhibition using MD
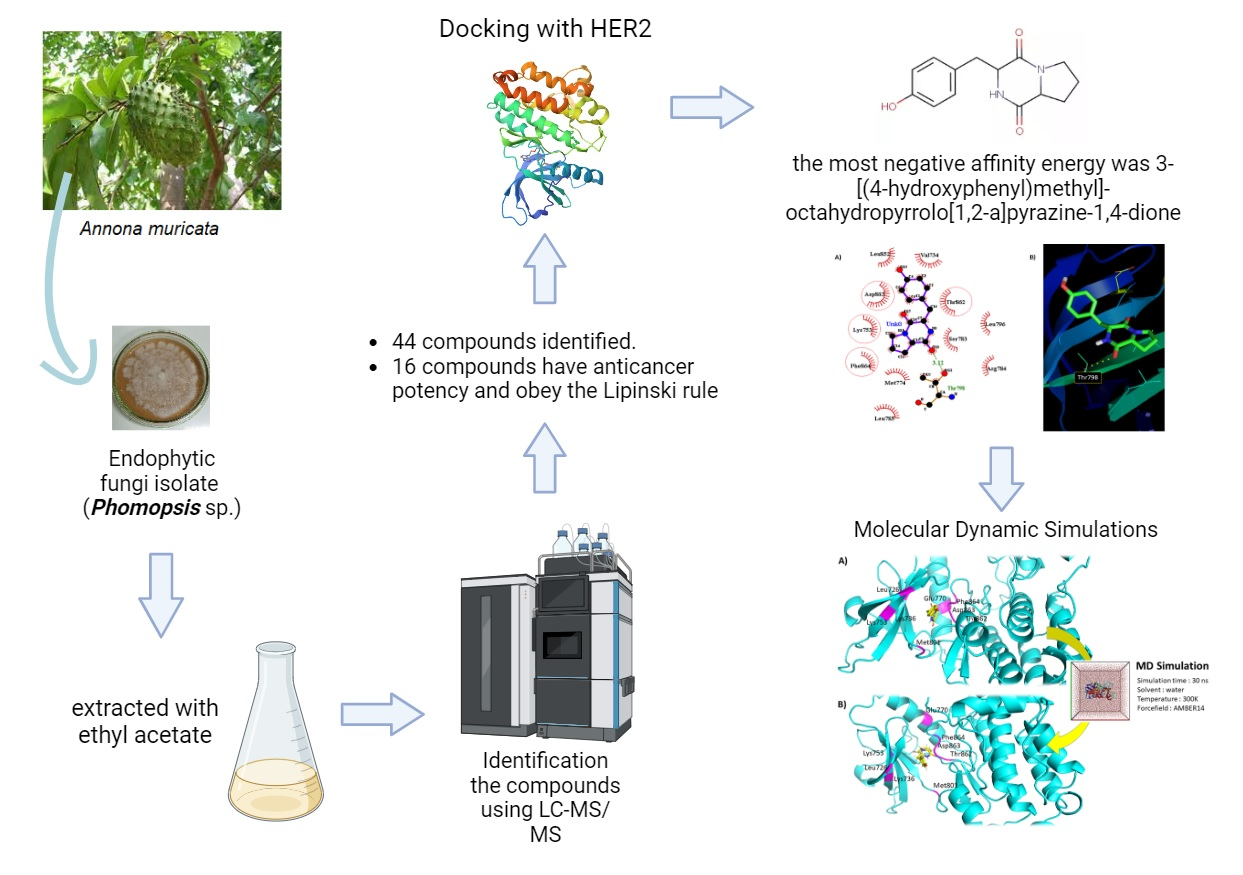
and MDS. Using LC-MS/MS, 44 compounds were successfully identified, and
16 have the potential to be anti-cancer and obey
Lipinski’s rule. In silico studies were
performed on the human epidermal growth factor receptor 2 (HER2).
Subsequently, molecular docking results showed that the most
negative affinity energy was
3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione (-9.4
kcal/mol), better than trastuzumab as a comparison ligand.
Molecular dynamic simulations (MDS) of protein-ligand complexes showed
prominent inhibition of HER2, as shown by dynamic trajectory analysis. Based on these
results, 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione
was identified as a promising HER2 inhibitor for breast cancer.
Breast cancer; Docking; HER2; Molecular dynamic; Phomopsis
Soursop leaves (Annona muricata) have been widely studied as medicinal plants that can be used as an alternative treatment for breast cancer (Dalal and Medithi, 2022). Using these leaves poses challenges as it demands precise harvesting timing and can negatively impact soursop fruit production, which is an important national agricultural commodity. Hence, the investigation of endophytic organisms in soursop leaves, known for their anticancer potential, becomes crucial. These symbiotic microorganisms, residing within host plants, can be isolated using plant growth media (Gouda et al., 2016). Endophytic organisms are closely related to their hosts, resulting in the transfer of genetic material and the production of secondary metabolites that are the same as their hosts (Kusari, Singh, and Jayabaskaran, 2014). Previous studies have shown that endophytic organisms in soursop leaves with the best anticancer potential are fungi, and the best solvent is ethyl acetate. According to ITS analysis, the type of endophytic fungi found in Indonesian Annona muricata leaves with anti-breast cancer potential belongs to Phomopsis sp. category (Minarni et al., 2017).
In vitro studies indicate that the ethyl
acetate extract from Phomopsis sp. shows a cytotoxic impact on breast
cancer cells. It effectively inhibits MCF-7 cell proliferation with an IC50
below 20 ppm, while demonstrating safety for normal cells (Minarni et al., 2017). In vivo
studies have also shown that Phomopsis extract at a dose of 20 mg/kgBW significantly decreased
the number and volume of breast tumors in DMBA-induced Sprague-Dawley
rats compared to the negative control group (Asyura et al., 2017). Molecular mechanism by which this extract can improve
breast cancer is not yet known.
Cancer occurs due to the abnormal proliferation of cells
in the body, resulting in uncontrolled cell growth (Maman and Witz, 2018). To prevent
the development of cancer cells, therapy is directed at inhibiting the
receptors that play a role in inhibiting the cell cycle and cancer cell
proliferation (Jazieh et al., 2020; Mutebi et al.,
2020).
Subsequently, several proteins play a role in breast cancer, including the human
epidermal growth factor 2 receptor (HER2).
HER2 is a
transmembrane receptor tyrosine kinase, part of the EGFR family, facilitating
the rapid growth of breast cancer cells (Feng et al., 2018). In normal
cells, HER2 activation regulates processes
such as proliferation, motility, and survival through cell signaling pathways. However, in breast cancer, overexpression of HER2 occurs predominantly
through amplification of the HER2 gene. This
overexpression is associated with a more aggressive
phenotype and is also an important predictive biomarker of the response to
HER2-targeted therapies, such as trastuzumab (Yu et al., 2020). HER2
testing is recommended for all patients with invasive breast cancer to
determine appropriate treatment options.
Before performing in vivo molecular analysis, it
was necessary to determine the content of the ethyl acetate extract of Phomopsis.
Considering the involvement of
various proteins in breast cancer incidence, an initial in silico analysis is
imperative to predict the influence of compounds in this extract on specific
proteins. In this study, bioinformatics analysis was performed using
molecular docking (MD) and
molecular dynamic simulations (MDS). Subsequently, MD is a
computational method used in molecular modeling to predict the preferred
orientation of one molecule to another when a ligand and target are bound to
each other to form a stable complex (Kaur et al., 2019). Lipinski's Rule of Five was an important criterion in
screening compounds before docking, serving as a rule of thumb to evaluate
drug-likeness and assess the probability of chemical compounds becoming an oral drug in humans. Biologically active molecules must meet these five conditions for their
potential use as oral drugs. These rules are related to molecular properties
that are important for drug pharmacokinetics in the human body, such as
absorption, distribution, metabolism, and excretion (ADME) (Benet et al., 2016).
MDS is a computer simulations method used in the
theoretical study of biological molecules, such as proteins and nucleic acids,
to analyze the physical movement of their constituent atoms and molecules over
time. This method has been applied to thoroughly examine dynamic of biological molecules,
their complexes, and their conformational changes by providing detailed
information about their fluctuations and conformational changes (Badar et
al. 2022). Furthermore, it is often used to study
protein-ligand docking interactions in the search for new drug candidates. In
MDS, atoms and molecules are allowed to interact for a fixed period, thereby
providing a view of dynamic "evolution" of a system (Liu et al., 2018).
Simulations can be used to study various properties of a system, such as its
thermodynamic, transport, and structural properties.
In silico studies on Phomopsis exist, but have not been
associated with breast cancer. Several studies have
investigated anticancer activity of Phomopsis, but they differ in observed proteins and Phomopsis sources, often
derived from different plant endophytes with distinct active compounds.
Therefore, this study aimed to identify the active
compounds in Phomopsis ethyl acetate extract and predict molecular
mechanism of action of these compounds against HER2 through MD and MDS.
The entire series of studies were conducted in
Bogor,
Indonesia. Extraction process was carried out at BRIN Cibinong. LCMS and in silico study was carried out at IPB University,
2.1. Phomopsis Extraction and Identification Chemical Compounds
In this study, the analytical material used was Phomopsis sp.
isolate (endophytic fungi from Indonesian Annona muricata leaves) that was obtained from BRIN
Indonesian Culture Collection (InaCC), labeled "Sir-G5". Phomopsis was first inoculated in Yeast Malt Agar (YMA) and then
cultivated in Yeast Malt Broth (YMB) for 21 days. The culture was then
extracted with ethyl acetate for 24 h using the maceration method and dried in a rotary evaporator to remove the solvent (Figure S1) (Minarni et al., 2017). Additionally, chemical compounds were identified in Phomopsis
extract using LC-MS/MS Thermo Scientific Vanquish Flex Ultra High-Performance
Liquid Chromatography (UHPLC) tandem Q Exactive Plus Orbitrap High-Resolution
Mass Spectrometer (Saravanakumar et al.,
2021).
2.2. Constructing Database of Phomopsis Extract Chemical Compounds
The resulting compounds from
LC-MS/MS analysis were then searched for their 2D structures. Databases
constructing the 2D structures were collected from PubChem (https://pubchem.ncbi.nlm.nih.gov) and ChemSpider (http://www.chemspider.com) in the SDF format (Kim et al., 2019). A 2D structure is required for the druggability analysis
of each chemical compounds. Drugability was predicted using the SwissADME database (http://www.swissadme.ch) and bioavailability prediction by Lipinski's "rules
of five" (Daina, Michielin, and
Zoete, 2017). Ligands showed
high bioavailability potential when they adhere to Lipinski's rules, which include molecular weight <500 Da, log P <5, H-bond donors
<5, H-bond acceptors <10, and molar refraction 40-130) (Chagas,
Moss, S., Alisaraie, 2018). These rules serve as a parameter for assessing
drug bioavailability,
specifically related to
the ADME properties of a drug.
2.3. Ligand and Protein Preparation for MD
The 2D structure was converted to 3D and saved in PDB
format for MD analysis. The 3D structures of Phomopsis compounds acted as
ligands (file type. pdb). Ligand optimization was performed using AutoDock Tools
1.5.6, by adjusting the torsion ligands and were saved in the
PDBQT format (Sahlan et al.,
2023). The protein
used in this in silico study was
human HER2 (PDB: 3PP0). The
data were downloaded from the PDB database (Protein Data Bank)
(http://www.rscb.org/pdb). Additionally, MD was performed using AutoDock Vina version 1.2.3. All data were processed using Intel Pentium Core i7
hardware (16 GB RAM, Windows 10, 64-bit). The HER2 receptor in PDB format was prepared using the
Discovery Studio Visualizer by removing water molecules and other ligands
attached to their structure. Hydrogen atoms were added using AutoDock Tools 1.5.6, and
the files were saved in PDBQT format (Fitrilia et al.,
2020).
Grid-box
validation was performed with a target root-mean-square deviation (RMSD) value
of less than 2 Å. The selected ligands were subjected to MD using the AutoDock Vina application by being attached to the
receptor target. Docking
results were scored and the best affinity energy was determined based on the
most negative value. The
ligand's binding area to the target receptor was identified, and the selected
ligand underwent MDS using YASARA software.
2.4. MDS
Phomopsis
chemical compounds exhibited strong affinity during docking on HER2, with the
most robust interaction observed for a specific ligand, showed by the most
negative value. The selected ligand was then analyzed for its interaction stability
with HER2
through MDS using YASARA Structure version 19.9.17 with the AMBER14 force
field (Prasasty and Istyastono, 2020; Bhadra and Siu, 2019). The
cell extension on each side around the solute was measured at 10 Å from the
cube box wall with periodic boundary conditions. MD simulations were performed
for 30 ns. The stability of the ligand-protein complex interaction was observed
based on the RMSD of the ligand and RMS fluctuations (RMSF).
3.1. Chemical Compounds of Phomopsis Extract
Analysis of Phomopsis sp. extract using LC-MS/MS
successfully identified 44 chemical compounds (Table S1). A review of the literature on the 44 compounds
identified in Phomopsis extract shows their potential applications as
antibacterial, anticancer, antioxidant, anti-inflammatory, and antimicrobial
agents, and as raw materials for industrial purposes (Figure S3). All the compounds had molecular weight of less than 500 Da.
Based on a literature review, there were 16 compounds with anticancer activity,
and all complied with more than three of Lipinski's rules (Table 1). These 16
compounds were subjected to molecular docking (MD) analysis.
Table 1 ADME properties of selected compounds conform to Lipinski's rules
|
Compounds |
Molecular Weight (Da) |
H-bond donor |
H-bond acceptors |
log p |
Molar refractivity |
|
1.
7-Hydroxycoumarine |
162.140 |
1 |
3 |
1.32 |
42.776 |
|
2.
Sorbic acid |
112.130 |
1 |
2 |
0.48 |
27.377 |
|
3.
Cyclo(phenylalanyl-prolyl) |
244.120 |
1 |
4 |
0.72 |
66.811 |
|
4.
3-[(4-hydroxyphenyl)methyl]-octa hydropyrrolo[1,2-a]pyrazine-1,4-dione |
260.116 |
0 |
4 |
-1.4 |
60.945 |
|
5.
1,3,7-Trihydroxy-6-methoxy-4,5-diisoprenylxanthone |
410.173 |
3 |
6 |
5.16 |
114.206 |
|
6.
4-Methoxychalcone |
238.099 |
0 |
2 |
3.6 |
72.800 |
|
7.
Dibenzoylmethane |
224.084 |
0 |
2 |
3.14 |
66.162 |
|
8.
4-(hydroxymethyl)benzoic acid |
152.047 |
2 |
3 |
0.88 |
39.324 |
|
9.
Citral |
152.120 |
0 |
1 |
2.32 |
50.465 |
|
10. 9-Oxo-10(E),12(E)-octadecadienoic
acid |
294.400 |
1 |
3 |
5.06 |
87.384 |
|
11. 3-[(1-Carboxyvinyl)oxy]benzoic
acid |
208.037 |
2
|
5 |
1.36 |
50.805 |
|
12. (+)-ar-Turmerone |
216.320 |
0 |
1 |
3.44 |
71.076 |
|
13. 3-Allyl-2-hydroxybenzoic
acid |
178.062 |
2
|
3 |
1.81 |
48.967 |
|
Compounds |
Molecular Weight (Da) |
H-bond donor |
H-bond acceptors |
log p |
Molar refractivity |
|
14. Ferulic
acid |
194.058 |
2 |
4 |
1.50 |
51.329 |
|
15. Chalcone |
208.089 |
0 |
1 |
3.58 |
66.248 |
|
16. Hydroxycinnamic
acid |
150.067 |
1 |
2 |
1,70 |
42.399 |
Extraction
process was carried out using ethyl acetate, a rarely used solvent. The process
generally uses harmless solvents such as distilled water (Sulistiawati et al., 2023). However, according to this study, most
endophytic fungi were extracted using this solvent, as well as in previous
investigations (Minarni et al., 2017). The final extraction
process ensured that extract was free of ethyl acetate (Table S2).
3.2. MD and Energy
Affinity
The 16 selected chemical compounds were subjected to MD to the HER2
receptor (PDB 3PP0). Table 2
shows docking results, showing affinity energy values, inhibition constants, and binding site
similarity (BSS) for the assessed compounds. The compounds exhibiting the most favorable docking results
possessed the lowest affinity energy, approaching the values of the native and
comparative ligands. Affinity energy is widely used as a
determinant of "docking scores " (Sahlan et al., 2020).
Table 2 Energy affinity and BSS from
MD with HER2
|
Compounds |
Energy affinity (kcal/mol) |
Inhibition constants (µm) |
BSS (%) |
Hydrogen bond |
Hydrophobic interaction |
|
· Native
Ligand: 2-{2-[4-({5-chloro-6-[3-(trifluoromethyl)phenoxy]pyridin-3-yl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethoxy}ethanol |
-11.1
|
0.007 |
|
1 |
19 |
|
· Compare
Ligand: Trastuzumab |
-7.5 |
3.136 |
40 |
4 |
7 |
|
· 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione |
-9.4 |
0.126 |
55 |
1 |
11 |
|
· Chalcone |
-9.3 |
0.15 |
70 |
|
15 |
|
· 4-methoxy
chalcone |
-9.3 |
0.15 |
55 |
|
12 |
|
· 1,3,7-Trihydroxy-6-methoxy-4,5-diisoprenylxanthone
|
-9.3 |
0.15 |
55 |
2 |
13 |
|
· Dibenzoylmethane |
-8.9 |
0.294 |
55 |
|
11 |
|
· Cyclo(phenylalanyl-prolyl) |
-8.7 |
0.413 |
45 |
|
9 |
|
· (+)-ar-Turmerone |
-8.1 |
1.138 |
60 |
|
13 |
|
· 9-Oxo-10(E),12(E)-octadecadienoic acid |
-7.4 |
3.713 |
70 |
|
17 |
|
· 3-[(1-Carboxyvinyl)oxy]benzoic
acid |
-7.4 |
3.713 |
55 |
3 |
9 |
|
· 3-Allyl-2-hydroxybenzoic
acid |
-7.4 |
3.713 |
45 |
2 |
10 |
|
· Ferulic
acid |
-6.9 |
8.643 |
45 |
2 |
8 |
|
· 7-hydroxycoumarin |
-6.8 |
10.234 |
30 |
1 |
6 |
|
· Hydrocinnamic
acid |
-6.4 |
20.117 |
50 |
2 |
9 |
|
· 4-(hydroxymethyl)benzoic
acid |
-6.2 |
28.205 |
40 |
3 |
6 |
|
· Citral |
-6.1 |
33.397 |
45 |
|
10 |
|
· Sorbic acid |
-5.6 |
77.735 |
35 |
2 |
8 |
The compounds
3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione shows the
most negative affinity energy (-9.4 kkal/mol) for the HER2 receptor among Phomopsis
extract compounds, showing a robust affinity. It hydrophobically interacts with
four key amino acids in the active site: Thr862, Asp863, Phe864, and Lys753 (Figure 1). These four active sites also interact hydrophobically with the
native ligand. This compounds exhibit high lipophilicity and pharmacokinetic
properties, as showed by its negative LogP value (Table 1). The compound's high
lipophilicity suggests excellent interaction with hydrophobic environments,
enhancing its bioavailability. Molecules with increased lipophilicity typically
show improved permeability through the enterocyte phospholipid bilayer,
emphasizing the compound's potential efficacy.
Affinity energy is an important parameter in determining
the quality of MD results, signifying the strength of the interaction
between the ligand and the active site of the receptor (Du et al., 2016). The affinity energy, or change in Gibbs
free energy (), represents the driving force of all chemical reactions in
nature to measure the capacity of a system to perform maximum work at constant
temperature and pressure (Popovic and Minceva,
2020). Protein binding to a ligand occurs when the change in
affinity energy is negative. A more negative value signifies a spontaneous
reaction, showing a favorable and energetically favorable interaction between
the protein and ligand. The
affinity energy is directly proportional to the inhibition constant (Ki). The
Ki value can predict the ability of a compounds to inhibit its target protein (Muttaqin, 2019). A lower Ki value shows a better
inhibition ability.
Although the BSS value of this compounds was not as high
as that of chalcone, (+)-ar-Turmerone and 9-Oxo-10(E),12(E)-octadecadienoic
acid, it contained many hydrogen bonds and hydrophobic interactions. Hydrogen
bonds play an important role in protein folding, protein-ligand interactions,
and catalysis (Chen et al., 2016). The quantity and arrangement of
hydrogen bonds significantly influence the binding affinity between a ligand
and a receptor. Hydrophobic interactions, characterized by nonpolar molecules
in water, are essential for protein folding and also contribute to stabilizing
ligand binding to the receptor. Subsequently, these interactions play crucial
roles in determining the strength and stability of the overall ligand-receptor
complex (Bogunia and Makowski,
2020).
Figure 1 Visualization of the tested ligand (3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione) and HER2 interactions. (A)
Superimpose the tested ligand with the native and comparative ligand on HER2
(yellow = kinase domain; red = alpha-helix C; orange = catalytic loop; green =
activating loop; magenta stick = compound; cyan stick = native ligand; green
stick = comparative ligand); (B) visualization in 3D; and (C) visualization in
2D (red circles = HER2 active sites; dot-dot lines = hydrogen bond).
3.3. MDS
MDS was performed to identify the stability of the
protein-ligand complexes through dynamic trajectories. This process can be performed using various tools, such as YASARA, AMBER,
and graphical processing units (GPUs) (Prasasty and Istyastono, 2020; Suhartanto et
al., 2018). Some data can be obtained from MDS, including RMSD, RMSF,
solvent access for surface area (SASA), and radius of gyration (Rg).
The RMSD of liganded HER2 was calculated for the initial model over the 30 ns MDS period as shown in Figure 2. The stability of the protein-ligand complex was first assessed based on RMSD calculations, considering both ligand movement and conformation. The selected test ligand for MDS on HER2 was 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione. Based on the RMSD graph of ligand movement on HER2 (Figure 2A), the structure showed a short increase in the first 4 ns and a sharp increase from 5 to 10 ns, with an RMSD value of >3.0 Å. The ligand movement reached equilibrium over 30 ns with an RMSD value of approximately 2.0 Å. In contrast, the RMSD graph of ligand conformation showed conformational stability for 30 ns with an RMSD value of around 1.2-1.4 Å (Figure 2B). The binding of the selected ligand to HER2 did not significantly affect the conformational stability of HER2 during simulations time, as shown in the RMSD graph of HER2, which tended to be stable with an RMSD value of approximately 3.0 Å.
Figure 2 RMSD of 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione
on HER2: ligand
movement (A), ligand
conformation (B)
The RMSF profile comparative analysis of the
HER2 protein bound to the ligand molecule showed no
significant fluctuations in the HER2 catalytic site (Figure 3A). Although some regions within the protein-ligand complex showed a
moderately high degree of mobility with fluctuations ranging between 1 and 4 Å,
these were not considered significant for
this study because the main focus was on the
catalytic dynamic behavior.
SASA serves as a geometric measure of protein-surface
interactions in an external solvent environment. SASA value (nm2 or Å2) was
directly proportional to the proportion of amino acids in the protein exposed
to the solvent environment (Figure 3B). The
disruption of SASA alters the amino acids exposed to the solvent, consequently
affecting the overall conformation of the protein (Chen
and Panagiotopoulos, 2019). SASA
analysis of the HER2 protein-ligand complex showed that the values tended to be
stable, with an average area of 14000 Å2. This showed that the ligand in the HER2 catalytic
pocket did not cause an increase in solvent exposure to the protein surface. In
contrast, this ligand does not disrupt the conformation of HER2
protein folding.
The radius of gyration (Rg) serves as an indicator of conformational
equilibrium, reflecting the compaction of the protein structure through folding
and unfolding processes (Liu et al., 2017). Based on the calculation of the Rg value of the HER2
protein-ligand complex (Figure 3C), the Rg value did not show significant deviations. The
Rg values tended to stabilize at approximately 19.8 - 20.0 Å. This shows that the presence of the ligand in the
catalytic pocket of HER2 did not induce a substantial change in the
conformational equilibrium, particularly in protein folding.
Molecular
dynamic of the HER2 and the ligand complex over 30 ns are shown in Figure 4. Based
on dynamic trajectory analysis, the tested ligand showed good stability in binding to the HER2 catalytic
pocket. A greater stability of the ligand when docked to the target protein
implies a stronger binding affinity. This suggests that the ligand is more
effective in inhibiting or interfering with the catalytic activity of the
target protein.
The MD and MDS results showed that 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione has the potential to inhibit HER2, a protein that plays a role in the incidence of breast cancer. Given the low relative abundance of 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione in Phomopsis extract, additional steps are essential to increase its quantity. One method is isolating the target compounds, facilitating a more concentrated and effective use for potential applications. One of the advantages of natural extract is the synergy between their constituent compounds. In this study, the compounds that could inhibit HER2 was 3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione, but another compounds could interact with other proteins, such as Thymidine Kinase, p53, or cyclin-dependent kinase. Therefore, it is necessary to analyze other proteins involved in the incidence of breast cancer.
Figure 3 RMSF values (A),
SASA (B), and Rg (C) of HER2 that complexed with ligands in 30 ns
Figure
4 3D
visualization of HER2-ligand complex,
before (A) and after (B) MD simulations
LCMS analysis
successfully identified 44 chemical compounds in extract of Phomopsis
sp. 16 of which have the potential to be anticancer and meet Lipinski’s rules. Additionally, MD results showed that the most negative energy affinity
for HER2 receptors was
3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione (-9.4 kcal/mol). MDS of the
protein-ligand complex showed prominent
HER2 inhibition as shown
by dynamic trajectory analysis. The
compounds
3-[(4-hydroxyphenyl)methyl]-octahydropyrrolo[1,2-a]pyrazine-1,4-dione was
identified as HER2 inhibitor, which can be developed for breast cancer therapy.
This study
was funded by The Indonesia Endowment Funds for Education and the Center for
Higher Education Fund, Ministry of
Education, Culture, Research and Technology of the Republic of Indonesia.
| Filename | Description |
|---|---|
| R1-CE-6696-20230919235616.docx | --- |
| R1-CE-6696-20231129153404.pdf | --- |
Asyura, C.,
Hasan, A., Julistiono, H., Husnawati, H., Bermawie, N., Riyanti, E., 2017.
Effectiveness of Ethyl Acetate Extract of Endophytic Fungi in Soursop Leaves
Towards The Growth of Mammary Tumor Induced By 7,12-Dimethylbenz Anthracene
in Female Rats. Annual Research &
Review in Biology, Volume 2017, pp. 1–8
Badar, M.S., Shamsi, S., Ahmed, J., Alam, M.A. 2022. Molecular Dynamics
Simulations: Concept, Methods, and Applications. In: Rezaei, N. (eds) Transdisciplinarity: Integrated Science, Volume 5, pp.
131–151
Benet, L.Z.,
Hosey, C.M., Ursu, O., Oprea, T.I., 2016. BDDCS, the Rule of 5 and Drugability. Advanced Drug Delivery Reviews, Volume 101,
pp. 89–98
Bhadra, P., Siu,
S.W., 2019. Refined Empirical Force Field to Model Protein–Self-Assembled
Monolayer Interactions Based on AMBER14 and GAFF. Langmuir, Volume 35(29), pp. 9622–9633
Bogunia, M.,
Makowski, M., 2020. Influence of Ionic Strength on Hydrophobic Interactions in
Water: Dependence on Solute Size and Shape. The
Journal of Physical Chemistry B, Volume 124(46), pp. 10326–10336
Chagas, C.M., Moss,
S., Alisaraie, L., 2018. Drug metabolites and their effects on the development
of adverse reactions: Revisiting Lipinski’s Rule of Five. International Journal of Pharmaceutics, Volume 549(1-2), pp. 133–149
Chen, D.,
Oezguen, N., Urvil, P., Ferguson, C., Dann, S.M., Savidge, T.C., 2016.
Regulation of Protein-Ligand Binding Affinity by Hydrogen Bond Pairing. Science Advances, Volume 2(3), p. e1501240.
Chen, H.,
Panagiotopoulos, A.Z., 2019. Molecular Modeling of Surfactant Micellization
Using Solvent-Accessible Surface Area. Langmuir,
35(6): pp. 2443–2450
Daina, A.,
Michielin, O., Zoete, V., 2017. SwissADME: a Free Web Tool to Evaluate
Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small
Molecules. Scientific Reports, Volume
7(1), p. 42717
Dalal, D.,
Medithi, S., 2022. A Review on The Importance of Annona Muricata Crude Extract (AMCE) as a Nutraceutical
Anti-Metastatic and its Coping Mechanism Against Breast Cancer. Current Nutrition & Food Science, Volume
18(5), pp. 466–475
Du, X., Li, Y.,
Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., Liu, S.-Q., 2016.
Insights Into Protein–Ligand Interactions: Mechanisms, Models, and Methods. International Journal of Molecular Sciences,
Volume 17(2), p. 144
Feng, Y., Spezia,
M., Huang, S., Yuan, C., Zeng, Z., Zhang, L., Ji, X., Liu, W., Huang, B., Luo,
W., 2018. Breast Cancer Development and Progression: Risk Factors, Cancer Stem
Cells, Signaling Pathways, Genomics, and Molecular Pathogenesis. Genes & Diseases, Volume 5(2), pp. 77–106
Fitrilia, T.,
Kurniawan, M.F., Kurniawati, F.R., Setiawan, T., 2020. The Potential of
Butterfly Pea Flower Methanol Extract as an Antioxidant by in Silico. Indonesian Journal of Applied Research
(IJAR), Volume 1(3), pp. 163–169
Gouda, S., Das,
G., Sen, S.K., Shin, H.-S., Patra, J.K., 2016. Endophytes: A Treasure House of
Bioactive Compounds of Medicinal Importance. Frontiers in Microbiology, Volume 7, p. 1538
Jazieh, K.,
Bell, R., Agarwal, N., Abraham, J., 2020. Novel Targeted Therapies For
Metastatic Breast Cancer. Annals of
Translational Medicine, Volume 8(14): p.
907
Kaur, T.,
Madgulkar, A., Bhalekar, M., Asgaonkar, K., 2019. Molecular Docking in
Formulation and Development. Current Drug
Discovery Technologies, Volume 16(1), pp. 30–39
Kim, S., Chen,
J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B.A.,
Thiessen, P.A., Yu, B., Zaslavsky, L., Zhang, J., Bolton, E.E., 2019. PubChem
2019 Update: Improved Access to Chemical Data. Nucleic Acids Research, Volume 47(D1), pp. D1102–D1109
Kusari, S., Singh, S., Jayabaskaran, C., 2014. Rethinking
Production of Taxol®(Paclitaxel) using Endophyte Biotechnology. Trends in Biotechnology, Volume 32(6), pp.
304–311
Liu, P., Lu, J.,
Yu, H., Ren, N., Lockwood, F.E., Wang, Q.J., 2017. Lubricant Shear Thinning
Behavior Correlated With Variation of Radius of Gyration via Molecular Dynamics
Simulations. The Journal of Chemical Physics,
Volume 147(8), p. 084904.
Liu, X., Shi,
D., Zhou, S., Liu, H., Liu, H., Yao, X., 2018. Molecular Dynamics Simulations
And Novel Drug Discovery. Expert Opinion on
Drug Discovery, Volume 13(1), pp.
23–37
Maman, S., Witz,
I.P., 2018. A History of Exploring Cancer in Context. Nature Reviews Cancer, Volume 18(6), pp. 359–376
Minarni, Artika,
I.M., Julistiono, H., Bermawie, N., Riyanti, E.I., Hasim, Hasan, A.E.Z., 2017.
Anticancer Activity Test of Ethyl Acetate Extract of Endophytic Fungi Isolated from
Soursop Leaf (Annona Muricata L.). Asian Pacific Journal of Tropical Medicine,
Volume 10(6), pp. 566–571
Mutebi, M.,
Anderson, B.O., Duggan, C., Adebamowo, C., Agarwal, G., Ali, Z., Bird, P.,
Bourque, J.M., DeBoer, R., Gebrim, L.H., Masetti, R., Masood, S., Menon, M.,
Nakigudde, G., Ng'ang'a, A., Niyonzima, N., Rositch, A.F., , El Saghir, N.S.,
Gralow, J.R., Eniu, A. 2020. Breast Cancer Treatment: A Phased Approach to
Implementation. Cancer, Volume 126, pp. 2365–2378
Muttaqin, F.Z., 2019.
Molecular Docking and Molecular Dynamic Studies of Stilbene Derivative
Compounds as Sirtuin-3 (Sirt3) Histone Deacetylase Inhibitor on Melanoma Skin
Cancer and Their Toxicities Prediction. Journal
of Pharmacopolium, Volume 2(2), p. 489
Popovic, M.,
Minceva, M., 2020. A Thermodynamic Insight Into Viral Infections: Do Viruses in
a Lytic Cycle Hijack Cell Metabolism Due to Their Low Gibbs Energy? Heliyon, Volume 6(5), p. e03933
Prasasty, V.D.,
Istyastono, E.P., 2020. Structure-Based Design and Molecular Dynamics
Simulations of Pentapeptide AEYTR as a Potential Acetylcholinesterase
Inhibitor. Indonesian Journal of
Chemistry, Volume 20(4), pp. 953–959
Sahlan, M.,
Dewi, L.K., Pratami, D.K., Lischer, K., Hermansyah, H., 2023. In Silico Identification
of Propolis Compounds Potential as COVID-19 Drug Candidates Against SARS-CoV-2
Spike Protein. International Journal of Technology, Volume 14(2), pp.
387–398
Sahlan, M.,
Faris, M.N.H.A., Aditama, R., Lischer, K., Khayrani, A.C., Pratami, D.K., 2020.
Molecular Docking of South Sulawesi Propolis against Fructose
1,6-Bisphosphatase as a Type 2 Diabetes Mellitus Drug. International Journal
of Technology, Volume 11(5), pp. 910–920
Saravanakumar,
K., Park, S., Sathiyaseelan, A., Kim, K.-N., Cho, S.-H., Mariadoss, A.V.A.,
Wang, M.-H. 2021. Metabolite Profiling of Methanolic Extract of Gardenia
Jaminoides by LC-MS/MS and GC-MS and its Anti-Diabetic, and Anti-Oxidant
Activities. Pharmaceuticals, Volume 14(2),
p. 102
Suhartanto, H.,
Yanuar, A., Wibisono, A., Hermawan, D., Bustamam, A., 2018. The Performance of
a Molecular Dynamics Simulation for the Plasmodium falciparum Enoyl-acyl
carrier-protein Reductase Enzyme using Amber and GTX 780 and 970 Double
Graphical Processing Units. International Journal of Technology, Volume
9(1), pp. 150–158
Sulistiawati,
E., Rochmadi, R., Hidayat, M., Budiman, A., 2023. Enhancement of Phycocyanin
Extraction from Dry Spirulina platensis Powder by Freezing-Thawing
Pre-treatment. International Journal of Technology, Volume 14(4), pp. 780–790
Yu, L., Fu, F.,
Li, J., Huang, M., Zeng, B., Lin, Y., Mei, Q., Lv, J., Wang, C., 2020. Dual HER2
Blockade versus a Single Agent in Trastuzumab-Containing Regimens for HER2-Positive
Early Breast Cancer: A Systematic Review and Meta-Analysis of Randomized
Controlled Trials. Journal of Oncology,
Volume 2020, p. 5169278
