Detection of Atrial Fibrillation using a Feedforward Sequential Model
Corresponding email: jan_michael_santos@dlsu.edu.ph
Published at : 07 Dec 2023
Volume : IJtech
Vol 14, No 7 (2023)
DOI : https://doi.org/10.14716/ijtech.v14i7.6684
Santos, J.M., Anit, E., Ramos, C.M., Bugtai, N., Sy, A., Roxas, N., Munsayac, F., 2023. Detection of Atrial Fibrillation using a Feedforward Sequential Model. International Journal of Technology. Volume 14(7), pp. 1506-1516
| Jan Michael Santos | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Edison Anit | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Catherine Manuela Ramos | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines. 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Nilo Bugtai | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Armyn Sy | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Nicanor Roxas | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
| Francisco Munsayac | 1. Department of Manufacturing and Management Engineering, De La Salle University, 2401 Taft Avenue, Manila, Philippines, 2. Institute of Biomedical Engineering and Health Technologies (IBEHT), De La |
Atrial
Fibrillation (AFib) and its associated symptoms are significant problems that
doctors and several studies have attempted to solve throughout the years. It is
diagnosed by analyzing a patient’s electrocardiogram (ECG) data. However,
continuous efforts have been made to develop an algorithm that detects AFib
with optimal efficiency and cost-effectiveness. In this study, a sequential
model was used based on feedforward neural network as this is arguably the
simplest algorithm developed and requires minimal computing power. The results showed that training the algorithm for 1000 epochs yielded the
best results. Further studies showed that using a combination of 10-fold
cross-validation and blindfold validation proved an ideal way to determine the
model's capabilities in distinguishing patients with AFib from those without.
In conclusion, the developed model successfully distinguished between AFib and
non-AFib patients with a 96.67% sensitivity, 94.61% specificity, and 95.64%
accuracy.
Atrial fibrillation; ECG; Feedforward neural network; Sequential method
Arrhythmia is an irregularity in the heartbeat,
manifesting as an increase or decrease in the heart's speed (MedlinePlus, 2016).
The most common type of arrhythmia is atrial fibrillation (AFib). Patients with
AFib frequently overlook symptoms, unaware that they are manifesting signs of
potential heart problems. This leads to an estimated half of AFib patients
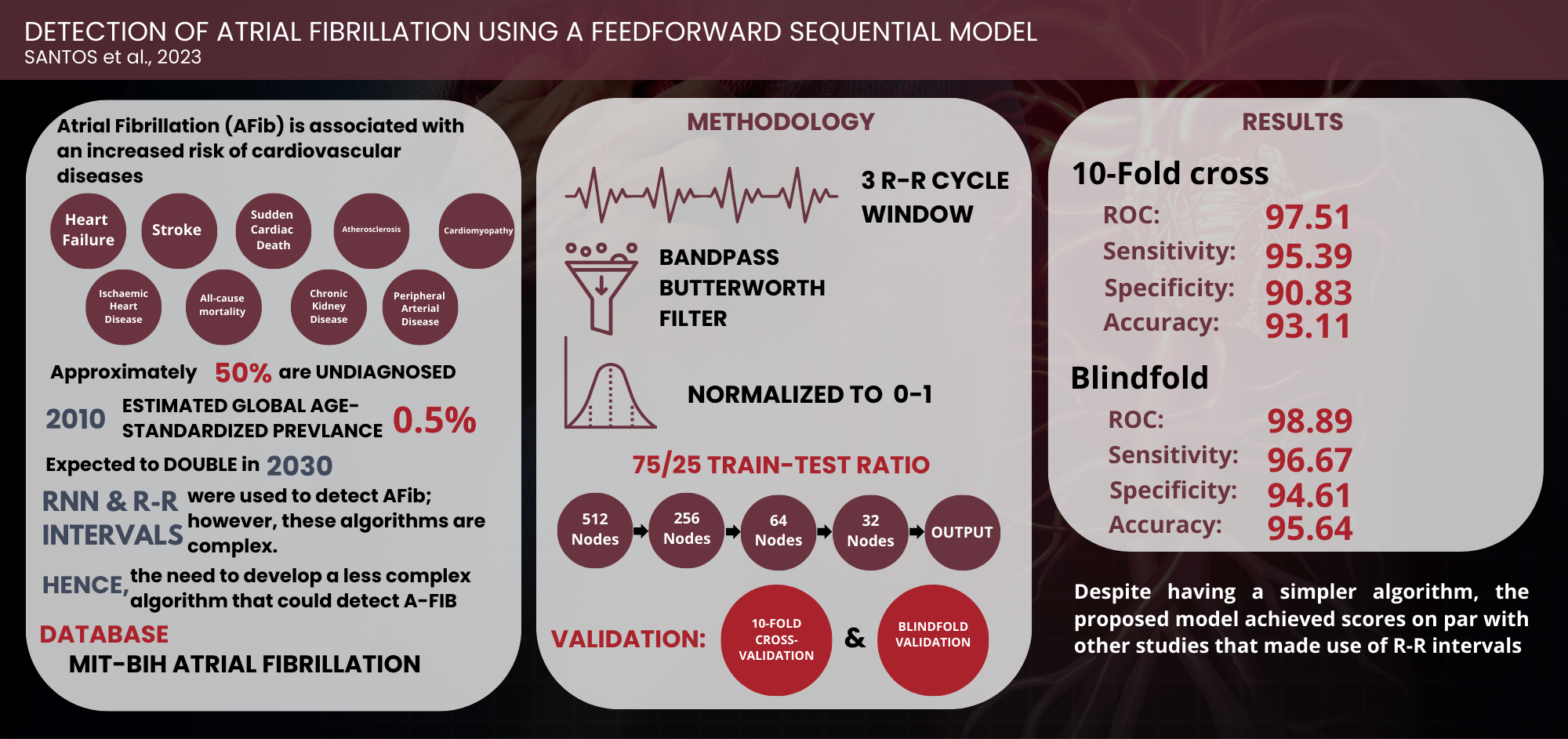
being undiagnosed (Atrial Fibrillation Association, 2012). In 2010, it had an estimated global age-standardized prevalence of
0.5%, with this expected to double by 2030 (Patel et al., 2018).
Afib has also been associated with an increased risk of numerous cardiovascular
conditions, such as heart failure, stroke, and sudden cardiac death (SCD) (National Health
Services, 2021; Ahmed and Zhu, 2020; Rattanawong et al., 2018; Odutayo et
al., 2016; Pistoia et al., 2016). Therefore,
early detection of AFib is necessary as it would lead to effective management
and improved patient outcomes (Hill et al., 2019).
EACTS’
2020 ESC Guidelines for the diagnosis and management of AFib (Hindricks et al.,
2020) state that the standard device used for detection
is the 12-lead ECG. However, for patients above 65 years, a cost-effective alternative
is pulse palpation. Taggar et al. (2015) reported a high false positive rate using pulse
palpation, with a sensitivity and specificity of 0.92 and
0.82, respectively, a positive likelihood ratio (PLR) of 5.2, and a negative
likelihood ratio (NLR) of 0.1. This suggests a need to develop an algorithm
that can accurately detect the presence of AFib while maintaining its
cost-effectiveness.
Several
studies have explored machine learning, with most model using Recurrent Neural
Network (RNN) and R-R intervals to detect AFib. However, RNN is a complex
algorithm and contrasts with Feedforward neural network, which is the most
straightforward algorithm devised (Poznyak, Oria, and Poznyak, 2019). In this
model, information is passed only in a single direction, moving from the input
nodes through the hidden nodes until it reaches the output nodes. The
simplicity of network makes it more manageable to operate and reduces the need
for robust and expensive devices for processing the data.
1.1. Atrial Fibrillation
One of the most common cardiac arrhythmias being treated is AFib. Arrhythmia refers to a change in the speed of the heartbeat. When a person has AFib, the atria experience irregular beating, causing poor blood circulation from the atria to the ventricles. It may occur in isolated incidents or be a chronic illness (Cai et al., 2020). The prevalence of AFib is increasing, with a 25% lifetime risk over the age of 40. Furthermore, complications include hemodynamic instability, cardiomyopathy, heart failure, and embolic events such as stroke.
Figure
1 Illustration of the P Wave, QRS
complex, and T wave signal of the ECG
Figure
1 shows an ECG waveform with AFib and a
waveform with a normal ECG, both taken from the dataset used in this study. The
P wave, QRS complex, and T wave constitute a normal waveform. In contrast, an
AFib waveform shows several inconsistent fibrillatory waves (F waves) replacing
the P wave, accompanied by R-R irregularities (irregular intervals between
successive R waves on the ECG). AFib can be diagnosed through a patient’s ECG
data, with features including [a] irregularly irregular rhythm, [b] absence of
P-wave, [c] variable ventricular rate, [d] QRS complexes usually less than
120ms, [e] fibrillatory waves that may be fine or coarse (amplitude of <0.5 mm
or >0.5 mm respectively), and [f] fibrillatory waves mimicking P-waves,
leading to misdiagnosis (Burns and Buttner, 2018). Deep
neural network (DNN) have gained
popularity for solving classification, segmentation, and detection issues.
Several deep learning algorithms have been used for AFib detection, including
the convolutional neural network (CNN), RNN, and autoencoder. With deep learning,
R-peak detection and removal of noises alleviate the burden of manual labor.
1.2. AI Algorithms
Episodes of AFib are often paroxysmal, requiring manual
diagnosis. Therefore, real-time cardiac monitoring with wearable health
trackers is required for the early detection of arbitrary events (Panindre, Gandhi,
and Kumar, 2020).
By using instantaneous heart rates (IHR) beat-to-beat variations of AFib could
be classified using the accuracy, sensitivity, specificity, precision, F1
score, recall, and area as criteria for evaluation and comparison.
Table 1 Comparison of performance of different supervised
learning algorithms
|
Algorithm |
Accuracy |
Precision |
Recall |
F1 Score |
AUC |
Specificity |
Training Duration | |
|
Logistic |
61.92 |
51.14 |
3.16 |
5.95 |
55.03 |
98.14 |
811 | |
|
AdaBoost |
61.60 |
49.03 |
17.47 |
25.76 |
63.66 |
88.80 |
46,504 | |
|
Gaussian |
62.90 |
52.13 |
33.26 |
40.61 |
65.96 |
81.17 |
235 | |
|
kNN |
74.99 |
79.24 |
46.82 |
58.86 |
77.56 |
92.41 |
1,456,010 | |
|
Decision Tree |
73.54 |
65.26 |
65.48 |
65.37 |
71.99 |
78.51 |
52,896 | |
|
Random |
81.89 |
79.15 |
71.30 |
75.02 |
90.35 |
88.42 |
3,129,625 | |
|
SVM - |
72.26 |
72.07 |
43.80 |
54.49 |
78.61 |
89.63 |
1,277,600 | |
|
LSTM |
67.20 |
56.49 |
61.11 |
58.68 |
75.39 |
70.95 |
9,663 | |
|
Bi-LSTM |
89.75 |
90.37 |
81.84 |
85.89 |
96.48 |
94.62 |
167,056 | |
As shown in
Table 1, several machine learning algorithms were trained and tested with a
dataset in PhysioNet using an NVIDIA Tesla V100 GPU of 32 GB memory. According
to the results, the Gaussian Naïve Bayes algorithm had the shortest training
duration, but its results were not promising compared to other algorithms. On
the contrary, the Bi-LSTM algorithm had the best performance among all the nine
tested algorithms. Additionally, Faust et al. (2018) applied
Bi-LSTM with R-R interval signals, achieving an accuracy of 98.51% and 99.77%
after 10-fold cross-validation and blindfold validations.
A possible reason for the superior performance of RNNs
over other network is their ability to overcome the critical impediments of
using standard machine learning algorithms. This includes the presumption that
inputs and outputs in model are independent of each other (Schmidhuber, 2015).
RNNs achieve this by permitting network to retain or use state data,
colloquially called "memory" which captures all input data.
Aside from RNNs, CNN are also used to detect AFib because
they require no manual feature extraction (Murat et al., 2021).
Reinforcement learning (RL) methods
have been applied to address challenges in traditional machine learning tasks,
particularly those emphasizing classification and the prediction process or
sequential processes such as budgeted classification and time prediction). In
addition, the evolution of deep architectures, or DNN from Neural Network (NN),
has expanded its applications, including but not limited to image
classification, audio recognition, machine translation, and natural language
processing. NN is a sequential decision process that chooses one mapping from a
group of candidate mappings at each layer of a deep design. On the other hand,
the Deep Sequential Neural Network (DSNN) model processes input through a
series of local rather than global transformations.
Denoyer and
Gallinari (2014) compared two alternative model, including (1)
NN or primary neural network and (2) DSNN-k or sequential model, where k is the
number of possible actions. The initial trials were conducted on five
University of California Irvine (UCI) datasets, which are low-dimensional
datasets with around 1,000 training samples. These results showed that using
more complex architectural designs did not improve the performance of models
for specific datasets (diabetes, heart). In these cases, a basic linear model
was adequate for computing results with high accuracy. Therefore, the DSNN
strategy performed better than the NN method,
particularly when the number of children per node is small.
1.3. Metrics
Diagnostic accuracy measurements include sensitivity,
specificity, predictive values, likelihood ratios, the area under the ROC
curve, Youden's index, and diagnostic odds ratio (Pennsylvania State University,
2013). This study focused on sensitivity and specificity,
which provide necessary measurements for patient screening. Sensitivity or True
Positive Rate (TPR) provides a measurement of how effectively models could
identify positive instances, while specificity or True Negative Rate (TNR)
measures the proportion of true negatives. A model with high specificity shows
that it could almost flawlessly identify the negative results. Ideally, models
should be highly sensitive and specific, but trade-offs occur between these
measurements as they are inversely proportional (Shreffler and Huecker, 2023).
A highly sensitive model captures most instances of positive results,
dismissing fewer cases of the disease. In screening applications, model should
achieve a higher specificity as it reduces false positives and minimizes
unnecessary diagnostic procedures for patients.
2.1. Model Setup
Figure 2 illustrates the proposed system architecture, consisting of the raw ECG data, an input layer with 512 nodes, three hidden layers of 256, 64, and 32 nodes, and a single output layer. Following the input layer, a 10% dropout was applied before passing through the first hidden layer to prevent overfitting. The size of the input layer was matched with the number of data points in a single window. This architecture is based on Feedforward neural network, where information passes through the layers once.
Figure
2 Proposed System Architecture
The dataset used was the MIT-BIH Afib Database (Goldberger et al., 2000), comprising 10-hour ECG recordings at 250 samples per second from 23 patients at Beth Israel Deaconess Medical Center. It provided rhythm annotations (.atr) for AFib, AFL (Atrial Flutter), J (AV junctional rhythm), and N (Normal) to represent all other rhythms. Figure 3 shows a sample of a 1-minute recording with rhythm annotations, processed with Python using the WFDB toolbox. Additionally, manually corrected beat annotation file (.qrsc) was used to detect the location of R-peaks. said it was observed that through these files, the different annotations could be visualized in the recordings, with N and AFib annotations presented in red and green, respectively.
Figure
3 A sample recording of manual
annotations and Python reading of raw data.
The
location of a record’s R-peaks was stored in an array. A random index (a random
R-peak) was then chosen, along with the next three indices to create a window,
as shown in Figure 4. Each window contained either N or AFib annotations, and
windows with different annotations were disregarded. Furthermore, the average
number of samples within a three-RR-cycle window was 544. This was resampled
for uniformity to 512, which is the nearest integer to the power of 2.
Figure
4 Illustration of the (a)
Windowed, (b) Filtered, and (c) Normalized Raw Data
A single window consists of three RR
cycles with a length of 512 data points. A Bandpass Filter with a low and high
pass cutoff frequency of 35Hz and 1Hz, respectively, was applied to the signals.
The amplitude of the signals was then scaled to return values between 0 and 1.
The signals were then arranged row-wise, with each window written to a single
row. AFib-annotated signals (Positive) were labelled as “1”, while those
annotated with N (Negative) were labelled as “0” before saving them separately
into a CSV file. The program adopted a 25% test and 75% train split.
Furthermore, multiple NumPy files (npz) were saved from the dataset for model
fitting and evaluation. The program, modelled with low computing power
applications in mind, was based on a Sequential Model with three layers,
excluding the input layer. The network
used an input layer of size 256, two hidden layers with sizes 64 and 32, and an
output layer of size 1. Model was evaluated using 10-fold cross-validation and
blindfold validation.
The proposed model was trained using an
NVIDIA RTX 3060 GPU with 12GB memory. Each epoch took approximately 0.446 s to train,
with the number of epochs set to 1000 based on previous training results
showing a sudden drop in accuracy beyond this point.
3.1. Validation Results
The confusion matrix and the Receiver Operating
Characteristic (ROC) Curve of the 10-fold cross-validation are shown in Figure
5. The ROC curve measures the model’s ability to correctly distinguish classes.
The closer the value is to 1, the better it can distinguish between classes. A
summary of the results can be seen in Table 2. Based on the cross-validation
results, the model returned a 0.9751 ROC curve, signifying that it accurately
distinguished the two classes. Additionally, the model exhibited higher
sensitivity than specificity, implying that it can better predict patients with
a disease than without.
Figure 5 Confusion
Matrix and ROC Curve of the (a) 10-fold and (b) blindfold validation
Table 2 Summary
of 10-Fold Cross-Validation and Blindfold Validation Performance
|
Validation Type |
TP |
TN |
FP |
FN |
Sensitivity |
Specificity |
Accuracy |
AUC |
|
10-Fold |
106,224 |
101,138 |
10,216 |
5,130 |
95.39% |
90.83% |
93.11% |
97.51 |
|
Blindfold |
107,650 |
105,357 |
5,997 |
3,704 |
96.67% |
94.61% |
95.64% |
98.89 |
3.2.
Discussion
Data is one of the most prominent
limiting factors in machine learning. Real-life applications of machine
learning model require a larger dataset than was used in this study. The
dataset was also balanced between AFib and non-AFib patients, which is not representative
of the real-world population. Therefore, the model is expected to have a bias
toward high sensitivity and false positives. Its performance can be compared to
other studies that used the same dataset, most of which were also cited in (Faust et al.,
2018), but with additional studies added.
Table 3 Performance
of another model using the same dataset
|
Author |
Data pre-processing |
Feature extraction method |
Analysis method |
Results |
|
Zhou et al., (2014) |
Median filter |
Shannon entropy |
Threshold evaluated with
ROC |
Sensitivity of 96.72%,
Specificity of 95.07%, |
|
Petrenas, Marozas, and Sörnmo (2015) |
8-beat sliding window |
Median filter and
threshold |
Threshold |
Sensitivity of 97.1%,
Specificity of 98.3% |
|
Henzel et al., (2017) |
Beat by beat evaluation, |
4 statistical features
and |
Generalized Linear Model |
Accuracy 93%, |
|
Faust et al., (2018) |
100 beat window, |
None |
Recurrent neural network |
Cross-validation and
Blindfold validation Accuracy: 98.51%, 99.77% |
|
Chen et al., (2022) |
Wavelet transform,
sliding window |
RR-interval |
Feedforward Neural
Network |
Cross-validation: Accuracy of 84%, |
|
Proposed
Model |
3
R-R cycle window, |
Annotations
provided |
Feedforward
Sequential Model |
Cross-validation
and Blindfold validation Accuracy:
93.11%, 95.64%, |
Table 3 shows that the proposed model
used a different method for predicting AFib aside from R-R intervals. Although
this method achieved scores comparable with other studies using R-R intervals,
most models still performed better. This could be attributed to the heavy focus
on developing a simple model and an insufficient complexity in capturing the
relationship between the input and output variables. A similar study by Chen et al.
(2022) also used the Feedforward model and reported a
cross-validation accuracy of 84%, a sensitivity of 84.26%, and a specificity of
93.24%. Comparing the proposed model to Chen et al. (2022), it
performed better based on the metric scores. In terms of model complexity, the
proposed model, with a total of 149,889 parameters, had a training duration of
approximately 0.443s per epoch using the NVIDIA RTX 3060 GPU. Among the studies
listed in Table 5, only Zhou et al. (2014) and Faust et al.
(2018) provided data regarding complexity. Zhou et al.
(2014) reported a training duration of 1.445s per 24 hours
of data, yet it remained unclear whether this value corresponded to the
training duration per epoch or the entirety of the training. The study also
specified the use of an Intel Pentium(R) Dual-Core E5800 processor. Assuming
the training duration was per epoch, it will be completed in only 0.443
seconds, making the proposed model faster. However, their model would be faster
for a duration covering the entire training process. Faust et al. (2018) presented another
model using a high-performance NVIDIA Quadro m5000 GPU tailored for industrial
use with 343,301 parameters and a training duration of 215s per epoch (total of
80 epochs). In comparison, the proposed model had fewer parameters and a faster
training duration, attributed to the unidirectional flow of information within
the network. For replication purposes, the codes
and datasets used in this study can be accessed through the link below:
https://github.com/JanMichaelSantos/Detection-of-Atrial-Fibrillation-using-Feedforward-Sequential-Model.git.
In conclusion, AFib is associated with an elevated
risk of heart failure, stroke, SCD, and other heart-related diseases. Its global
age-standardized prevalence is expected to double by 2030. This can be
addressed through an early diagnostic system by analyzing a patient’s ECG
recording. Previous studies used various algorithms combined with R-R intervals
for early detection. However, these necessitated hefty hardware. One objective
of this study was to develop a cost-effective diagnostic system. Based on
related literature, it was identified that sequential model could be
implemented, using ECG features alongside R-R intervals to predict and
accurately classify AFib and non-AFib patients. Model underwent training for
1000 epochs with a 75:25 train-test ratio and was filtered using a bandpass
Butterworth with cutoff frequencies of 1-35 Hz. Upon evaluation, the 10-fold
cross-validation and the blindfold validation performance yielded a
95.39-96.67% sensitivity, 90.83-94.61% specificity, 93.11-95.64% accuracy, and
an AUC of 97.51-98.89%, respectively. Although other model developed showed
higher performance on these metrics, they used complex algorithms that
increased computation time. On the contrary, the proposed model had a simpler
algorithm, making it more practical for implementations on small wearable
devices with low computing power. It is
recommended that future studies explore increasing the complexity of sequential
model and use different datasets to gain a
more comprehensive understanding of the model’s
performance on a broader representation. However, in implementing these
recommendations, it is vital to consider the computing power of wearable devices.
The authors
are extremely grateful to the Department of Manufacturing Engineering and
Management of the Gokongwei College of Engineering – De La Salle University for
providing the materials and equipment used in this study and to the LAPARA
project of the Institute of Biomedical Engineering and Health Technologies
(IBEHT), funded by the Department of Science and Technology – Philippine
Council for Health Research and Development (DOST-PCHRD). The authors are also
grateful to Engr. Jesse Daniel Santos for his valuable support.
Ahmed, N., Zhu, Y., 2020. Early Detection of Atrial
Fibrillation Based on ECG Signals. Bioengineering, 7(1), p. 16
Atrial Fibrillation Association, 2012. The AF
Report Atrial fibrillation: Preventing A Stroke Crisis. Atrial Fibrillation
Association. Available online at: http://www.preventaf-strokecrisis.org/files /files/The%20AF%20Report%2014%20April%202012.pdf,
Accessed on February 15, 2022
Burns, E., Buttner, R., 2018. Atrial Fibrillation. Life in the Fast
Lane. Available online at: https://litfl.com/atrial-fibrillation-ecg-library/,
Accessed on October 28, 2022
Cai, W., Chen, Y., Guo, J., Han, B., Shi, Y., Ji,
L., Wang, J., Zhang, G., Luo, J., 2020.
Accurate Detection of Atrial Fibrillation from 12-Lead ECG Using Deep
Neural Network. Computers in Biology and Medicine, Volume 116, p. 103378
Chen, Y., Zhang, C., Liu, C., Wang, Y., Wan, X.,
2022. Atrial Fibrillation Detection Using a Feedforward Neural Network. Journal
of Medical and Biological Engineering, Volume 42(1), pp. 63–73
Denoyer, L., Gallinari, P., 2014. Deep Sequential
Neural Network. arXiv.org. Available online at https://arxiv.org/abs/1410.0510,
Accessed on December 07, 2022
Faust, O., Shenfield, A., Kareem, M., San, T.R.,
Fujita, H., Acharya, U.R., 2018. Automated Detection of Atrial Fibrillation
Using Long short-term Memory Network with RR Interval Signals. Computers in
Biology and Medicine, Volume 102, pp. 327–335
Goldberger, A.L., Nunes, A., Glass, L., Hausdorff,
J.M., Ivanov, P.Ch., Mark, R.G., Mietus, J.E., Moody, G.B., Peng, C.-K.,
Stanley, H.E., 2000. PhysioBank, PhysioToolkit, and PhysioNet: Components of A
New Research Resource for Complex Physiologic Signals. Circulation,
Volume 101(23), p. e215
Henzel, N., Wrobel, J., Horoba, K., 2017. Atrial
Fibrillation Episodes Detection Based on Classification of Heart Rate Derived
Features. In: 2017 MIXDES - 24th International Conference
Mixed Design of Integrated Circuits and Systems, pp. 571–576
Hill, N.R., Ayoubkhani, D., McEwan, P., Sugrue, D.,
Farooqui, U., Lister, S., Lumley, M.,
Bakhai, A., Cohen, A.T., O’Neill, M., Clifton, D.A., Gordon, J., 2019.
Predicting Atrial Fibrillation in Primary Care Using Machine Learning. PLoS
One, Volume 14(11), p. e0224582
Hindricks, G., Potpara, T., Dagres, N., Arbelo, E.,
Bax, J.J., Blomström-Lundqvist, C., Boriani, G., Castella, M., Dan, G.-A.,
Dilaveris, P.E., Fauchier, L., Filippatos, G., Kalman, J.M., La Meir, M., Lane,
D.A., Lebeau, J.-P., Lettino, M., Lip, G.Y.H., Pinto, F.J., Thomas, G.N.
(2020). 2020 ESC Guidelines for the Diagnosis and Management of Atrial
Fibrillation Developed in Collaboration with the European Association of
Cardio-Thoracic Surgery (EACTS). European Heart Journal, Volume 42(5), pp. 373–498
MedlinePlus, 2016. Arrhythmia. National Library of
Medicine (US). Available online at
https://medlineplus.gov/arrhythmia.html, Accessed on February 09, 2022
Murat, F., Sadak, F., Yildirim, O., Talo, M., Murat, E., Karabatak, M.,
Demir, Y., Tan, R.-S. Acharya, U.R., 2021. Review of Deep Learning-Based
Atrial Fibrillation Detection Studies. International Journal of
Environmental Research and Public Health, Volume 18(21), p. 11302
National Health Services, 2021. Causes - Atrial
Fibrillation. National Health Services (NHS). Available online at
https://www.nhs.uk/conditions/atrial-fibrillation/causes/, Accessed on February
15, 2022
Odutayo, A., Wong, C.X., Hsiao, A.J., Hopewell, S.,
Altman, D.G., Emdin, C.A., 2016. Atrial Fibrillation and Risks of
Cardiovascular Disease, Renal Disease, and Death: Systematic Review and
Meta-Analysis. BMJ, Volume 354, p. i4482
Panindre, P., Gandhi, V., Kumar, S., 2020.
Comparison of Performance of Artificial Intelligence Algorithms for Real-Time
Atrial Fibrillation Detection using Instantaneous Heart Rate. In: 2020
IEEE 17th International Conference on Smart Communities: Improving
Quality of Life Using ICT, IoT and AI (HONET), pp. 168–172
Patel, N., Atti, V., Mitrani, R.D., Viles-Gonzalez,
J.F., Goldberger, J.J., 2018. Global Rising Trends of Atrial Fibrillation: A
Major Public Health Concern. Heart, Volume 104(24), pp.1989–1990
Pennsylvania State University, 2013. 11.3 – Sensitivity,
Specificity, Positive Predictive Value, and Negative Predictive Value.
PennState. Available online at https://online.stat.psu.edu/stat507/lesson/11/11.3-0,
Accessed on October 27, 2022
Petrenas, A., Marozas, V., Sörnmo, L., 2015. Low-Complexity Detection of Atrial Fibrillation in Continuous Long-Term
Monitoring. Computers in Biology and Medicine, Volume 65, pp. 184–191
Pistoia, F., Sacco, S., Tiseo, C., Degan, D.,
Ornello, R., Carolei, A., 2016. The Epidemiology of Atrial Fibrillation and
Stroke. Cardiology Clinics, Volume 34(2), pp. 255–268
Poznyak, T., Oria, I.C., Poznyak, A.S., 2019.
Background on Dynamic Neural Networks. In: Ozonation and Biodegradation in
Environmental Engineering. Elsevier. pp.57–74
Rattanawong, P., Upala, S., Riangwiwat, T.,
Jaruvongvanich, V., Sanguankeo, A., Vutthikraivit, W., Chung, E.H., 2018.
Atrial Fibrillation is Associated with Sudden Cardiac Death: A Systematic
Review and Meta-Analysis. Journal of Interventional Cardiac
Electrophysiology, Volume (2), pp. 91–104
Schmidhuber, J., 2015. Deep Learning in Neural
Networks: An Overview. Neural Networks, Volume 61, pp. 85–117
Shreffler, J., Huecker, M.R., 2023. Diagnostic
Testing Accuracy: Sensitivity, Specificity Predictive Values and Likelihood
Ratios. Nih.gov. Available online at
https://www.ncbi.nlm.nih.gov/books/NBK557491/, Accessed on December 07, 2022
Taggar, J., Coleman, T., Lewis, S., Heneghan, C., Jones, M., 2015. Accuracy Of Methods for Detecting an Irregular Pulse and Suspected Atrial Fibrillation: A Systematic Review and Meta-Analysis. European Journal of Preventive Cardiology, Volume 23(12), pp. 1330–1338
Zhou, X., Ding, H., Ung, B., Pickwell-MacPherson, E., Zhang, Y.-T., 2014. Automatic Online Detection of Atrial Fibrillation Based on Symbolic Dynamics and Shannon Entropy. Biomedical Engineering Online, Volume 13(1), pp. 1–18
