Design of Specific and Efficient sgRNA for CRISPR/Cas9 System to Knockout Superoxide Dismutase 2 in Breast Cancer Stem Cells
Corresponding email: septelia.inawati@ui.ac.id
Published at : 05 Feb 2024
Volume : IJtech
Vol 15, No 2 (2024)
DOI : https://doi.org/10.14716/ijtech.v15i2.6680
Arumsari, S., Wanandi, S.I., Syahrani, R.A., Watanabe, Y., Mizuno, S., 2024. Design of Specific and Efficient sgRNA for CRISPR/Cas9 System to Knockout Superoxide Dismutase 2 in Breast Cancer Stem Cells. International Journal of Technology. Volume 15(2), pp. 353-363
| Sekar Arumsari | Molecular Biology and Proteomics Core Facilities, Indonesian Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia, Jakarta 10430, Indonesia |
| Septelia Inawati Wanandi | 1. Molecular Biology and Proteomics Core Facilities, Indonesian Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia, Jakarta 10430, Indonesia, 2. Department of Biochem |
| Resda Akhra Syahrani | 1. Molecular Biology and Proteomics Core Facilities, Indonesian Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia, Jakarta 10430, Indonesia, 2. Doctoral Program in B |
| Yukihide Watanabe | Department of Experimental Pathology, Graduate School of Comprehensive Human Sciences, Faculty of Medicine, University of Tsukuba, Tsukuba 305-8575, Japan |
| Seiya Mizuno | Laboratory Animal Resource Center and Trans-border Medical Research Center, Faculty of Medicine, University of Tsukuba, Tsukuba 305-8575, Japan |
This study aimed to design specific and efficient single guide RNA
(sgRNA) for CRISPR/Cas9 system to knockout human superoxide dismutase 2 (SOD2)
in human breast cancer stem cells (BCSCs). To achieve this, two sgRNA targets
were selected, located within the Ala16Val polymorphism and the conserved
region of human SOD2 variants. Design process was carried out using
CRISPRdirect tool, considering the on/off target efficiency score.
Subsequently, these sgRNAs were cloned into CRISPR/Cas9 expression plasmid and transfected
into both the CD24-/CD44+ and ALDH1+ human BCSCs. To determine the most
efficient sgRNA, a cleavage activity assay was conducted. The effectiveness of
CRISPR/Cas9 system in knocking out the mRNA and protein expression of SOD2 in
BCSCs was determined using quantitative reverse transcriptase polymerase chain
reaction and western blot assays, respectively. The results showed that the
sodex2.1 sgRNA targeting the Ala16Val region within exon 2 was ineffective in
knocking out the SOD2 protein expression. However, among the four sgRNAs
targeting the conserved region of nine SOD2 variants, the sodex2.4 sgRNA
spanning from nucleotide 532-554 showed the highest efficiency based on
cleavage activity assays. The sodex2.4 sgRNA significantly decreased both mRNA
and protein expressions of SOD2 in human BCSCs. In conclusion, this study
successfully designed specific and efficient sgRNA to knockout SOD2 expression
in human BCSCs using CRISPR/Cas9 system. Moreover, further investigations are
recommended to understand the impact of SOD2 knockout on the aggressiveness of
breast cancer, particularly in BCSCs.
Breast cancer; CRISPR/Cas9; sgRNA; SOD2; Stem cells
Tissue engineering is currently playing a significant role in the rapid
progression of various technologies in medical applications (Irsyad et al., 2022). The integration of
gene editing strategies into tissue engineering, necessitating nuclease
modifications is essential
Cancer is a
multifaceted disease characterized by the unregulated growth of abnormal cells,
originating from cancer stem cells (CSCs). These CSCs form a side population
with stemness properties similar to normal stem cells, possessing high
tumorigenicity which contributes to the progression of cancer (Ahmad, Zain, and Aziz, 2018; Ayob and Ramasamy, 2018). Among all cases, breast
cancer is the most frequently diagnosed type, ranking as the first leading
cause of cancer-related death in women globally (Nusantara
et al., 2016). Previous studies have identified several CSC
populations in human breast cancer, including CD24-/CD44+ and aldehyde
dehydrogenase-1 (ALDH1+) cells (Shiraishi et
al., 2017; Wanandi et al., 2017).
The presence of breast cancer stem cells (BCSCs) strongly correlates with
biological aggressiveness, resulting in a poorer prognosis (Sarkar et al., 2018).
Several studies
have reported that the aggressiveness of BCSCs is closely correlated with the
overexpression of superoxide dismutase 2 (SOD2), a mitochondrial antioxidant
enzyme. SOD2 plays a critical role in safeguarding cells from excessive
superoxide, the primary reactive oxygen species (ROS) produced within the
mitochondria of cells (Azadmanesh, Trickel, and
Borgstahl, 2017). Initially regarded as a tumor suppressor, decreased
SOD2 expression was associated with cellular transformation and tumorigenesis
due to high ROS-mediated DNA damage, resulting from the accumulation of
superoxide and other oxidants. However, recent research has shown that the
overexpression of SOD2 in cancer cells induces a moderate increase in
sub-lethal H2O2, triggering oxidation and enhancing redox
signalling. This phenomenon promotes cells proliferation and increases cancer
aggressiveness, showing the dual role of SOD2 in tumorigenesis (Kim et al., 2017).
The SOD2 gene is
located on chromosome 6q35, comprising 5 exons and 4 introns. In this gene, a
well-studied single nucleotide polymorphism (SNP) known as Ala16Val in exon 2
causes changes in the 16th amino acid from valine to alanine. This SNP has been
correlated with the changes in SOD2 conformation, activity in mitochondria, and
risk of various diseases, including breast cancer (Sari
et al., 2019; Wang et al., 2018; Abdelrauf,
et al., 2017).
CRISPR/Cas9 gene editing technology has been applied to knockout SOD2 expression in HEK293T using sgRNA targeting exon 3 of the SOD2 gene (Cramer-Morales et al., 2015). The impact of this technology is limited to specific SOD2 variants that possess an intact exon 3. However, there is a lack of data regarding specific SOD2 variants within human BCSCs contributing to their aggressiveness. Consequently, this study aimed to design specific and efficient sgRNA for CRISPR/Cas9 system, targeting the Ala16Val SNP and the conserved region common to all SOD2 variants to knockout SOD2 expression in breast cancer cells, particularly the most aggressive BCSCs.
2.1. Cells Culture
ALDH1+ and CD24-/CD44+ BCSCs were obtained
from Cells Culture Laboratory for Cancer Stem Cells, Department of Biochemistry
and Molecular Biology, Faculty of Medicine, Universitas Indonesia. These cells
were cultured in a serum-free DMEM/F12 medium (Gibco, Thermo Fisher Scientific
Inc., Massachusetts, USA). Furthermore, the HEK293T cells line, derived from a
human embryonic kidney cells line, was sourced from ATCC or Riken Bioresource
Research Center (Wardhani et al., 2020;
Amalia et al., 2019) and maintained in DMEM (Gibco™, Thermo
Fisher Scientific Inc., Massachusetts, USA) supplemented with 10% Fetal Bovine
Serum (Gibco™, Thermo Fisher Scientific Inc., Massachusetts, USA), penicillin G
(Gibco™, Thermo Fisher Scientific Inc., Massachusetts, USA), streptomycin
sulfate (Gibco™, Thermo Fisher Scientific Inc., Massachusetts, USA), and 10
µg/mL of insulin. All cells line were incubated at 37°C with 5% CO2
and 20% O2.
2.2. CRISPR/Cas9 Genome Editing
This study has been granted ethical approval by the Ethics Committee of the Faculty of Medicine, University of Indonesia - Cipto Mangunkusumo Hospital, with number KET-B64/UN.2.F1/ETIK/PPM.00.02/2022. The schematic process of CRISPR/Cas9 genome editing of the SOD2 gene is illustrated in Figure 1.
Figure 1 Flowchart of CRISPR/Cas9 genome
editing of the SOD2
2.2.1. sgRNA Design
The initial
design of sgRNA targeting SOD2 in exon 2 was carried out using CRISPRdirect
website computational tool (https://CRISPR.dbcls.jp/)(Naito
et al., 2014) based on the NCBI database (NG_007829.4) (NCBI, 2023).
2.2.2. Construction of
CRISPR/Cas9 Plasmid
Designed
sgRNA sequences were inserted into CRISPR/Cas9 cloning plasmid
pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene, plasmid #62988, Watertown, USA)
using BbsI restriction sites. These sgRNA sequences were introduced into competent
E. coli DH5 cells through 1 mg/mL ampicillin selection. Bacterial
cultures were grown on LB agar plates at 37°C, followed by growth in LB broth
within a shaking incubator at 37°C. Subsequently, plasmids were extracted using
a High-speed Plasmid Mini Kit (Geneaid Biotech Ltd., New Taipei City, Taiwan).
2.2.3. Cleavage Efficiency Assay
Cleavage
efficiency of each sgRNAs was assessed using the enhanced green fluorescent
protein (EGFP) expression plasmid (pCAG-EGxxFP) (Addgene, plasmid #50716,
Watertown, USA) in HEK293Tcells, as previously described (Wardhani et al., 2020). The SOD2 target
sequences were ligated between EGFP fragments of pCAG-EGxxFP plasmid and
transformed into E. coli DH5a competent following the protocol of the
manufacturer. The resulting target plasmid was purified using a Miniprep
purification kit (Qiagen Ltd., Manchester, UK). Subsequently, the purified
plasmid was transfected with CRISPR/Cas9 plasmid expressing sgRNA SOD2 gene and
hCas9 into human cells using polyethylenimine (Sigma-Aldrich Chemie GmbH,
Taufkirkchen, Germany). The EGFP in transfected cells was observed with a
fluorescence microscope (Eclipse Ni-U with Intenslight, Nikon Instruments Inc.,
New York, USA). Quantification of fluorescence intensity was measured by ImageJ
version 1.53, compared to negative plasmid control without sgRNA expression
vector.
2.2.4. Preparation of Total RNA
and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Cells were
extracted using Tripure Isolation Reagent (Roche Applied Science, Basel,
Switzerland) based on the guidelines of the manufacturer, while concentrations
were quantified with a spectrophotometer. For qRT-PCR, SensiFAST™ SYBR® No-ROX
One-Step Kit (Bioline, Meridian Bioscience, London, UK) was conducted in a
Real-Time PCR System 7500 Fast Thermal Cycler (Applied Biosystems, Life
Technologies, California, USA), according to the protocol of the manufacturer.
Primer sequences for 18S rRNA were 5’-AAACGGCTACCACATCCAAG-3’ (forward) and
5’-CCTCCAATGGATCCTCGTTA-3’ (reverse), while SOD2 consisted of
5’-GCACTAGCAGCATGTTGAGC-3’ (forward) and 5’-ACTTCTCCTCGGTGACGTTC-3’ (reverse).
Each reaction was performed in triplicate and the relative mRNA expression
levels were determined using the Livak formula (Livak
and Schmittgen, 2001).
2.2.5. Western Blot Analysis of
SOD2
Total
protein was extracted using RIPA Lysis Buffer (Abcam, Cambridge, UK) and
protein concentration was measured using Bradford Assay (Bio-Rad Laboratories
Inc., California, USA). Equal amounts of protein (20 µg/lane) were loaded onto
15% SDS-PAGE gels and transferred to nitrocellulose membranes. Primary
antibodies used included rabbit anti-SOD2 1:1000 (Cells Signalling
Technologies, Massachusetts, USA) and mouse antiactin 1:2000 (Cells
Signalling Technologies, Massachusetts, USA). To enhance visualization,
chemiluminescence reagent (Abcam, Cambridge, UK) was used, while the blots were
captured using Chemiluminescence Documentation System (UVITEC Ltd., Cambridge,
UK).
2.3. Statistical analysis
All data presented were expressed as
means ± standard deviation from three independent experiments. Knockout data
were compared to WT (wildtype) cells without genome editing, and statistical
analysis was performed using an independent t-test.
SOD2 has been reported
to play dual and contradictory roles in tumorigenesis, acting as a tumor
suppressor by mitigating oxidative stress in the early stage of carcinogenesis.
Furthermore, it serves as a tumor promotor that stimulates cells proliferation
and metastasis by suppressing ROS-induced cytotoxicity in malignant tumors (Wanandi et al., 2017). In a previous
study, Morales et al. used CRISPR/Cas9 genome editing to target SOD2 in HEK293T
cells with sgRNA located in exon 3. Consequently, cells lacking SOD2 showed
reduced clonogenic potential due to impaired mitochondrial function resulting
from increased oxidative stress (Cramer-Morales et
al., 2015). Although SOD2 knockout studies have been carried out in
human cells, there is no report on cancer cells, particularly in BCSCs.
Several studies have showed that BCSCs show higher levels of
SOD2 compared to the non-BCSC population. This high expression strongly
correlates with their aggressiveness, including stemness and metastasis (Srivastava et al., 2023; Wanandi et al., 2019). However, specific
sequence variation in human SOD2 contributing to the aggressive properties
remains unknown. Previous investigations have shown that the Val/Val genotype
shows low SOD2 activity. Meanwhile, the Ala/Ala genotype, which expresses a
high SOD2 level, is significantly associated with increased cancer risk (Da-Cruz-Jung et
al., 2020; Wang et al., 2018). This SNP is not
present in any of the nine SOD2 variants published in the NCBI database (NCBI, 2023). Consequently, to knock out the SOD2 gene
in human BCSCs using CRISPR/Cas9 system, two sgRNA targets were used in this
study. These included one sgRNA target located within the Ala16Val polymorphism
region within exon 2 and the other in the conserved region common to all SOD2
variants.
In
this study, sgRNAs were designed using CRISPRdirect that can precisely identify
on-target locations, while minimizing the possibility of off-target effects (Karlapudi et al., 2018; Naito et al., 2014). The selected sgRNA has
a single exact match with the target sequence, specifically designed for
editing SOD2 in BCSCs. To target the
Ala16Val site at nucleotide (nt) 401-403, sgRNA was designed spanning from nt
385-407 within exon 2 of the SOD2 gene, as presented in Figure 2A. Before
transfecting CRISPR/Cas9 plasmid containing the sodex2.1 sgRNA into BCSCs, its
efficiency was assessed in a cleavage activity assay using HEK293T cells
transfected with a constructed pCAG-EGxxFP expression plasmid. The result
showed a moderate green fluorescence intensity produced from EGFP expression,
as presented in Figure 2B. This suggested moderate efficiency of sgRNA in
guiding Cas9 to cleave the DNA target. Subsequently, the effectiveness of the
sodex2.1 sgRNA for CRISPR/Cas9 system was evaluated by analyzing SOD2
expression in HEK293T cells, as depicted in Figure 2C. Although the relative
mRNA expression decreased to 0.6-fold, Cas9 cleavage had no significant impact.
This suggested that sgRNA targeting the Ala16Val SNP region was ineffective in
knocking out the SOD2 expression in HEK293T cells due to the absence of the
Ala16Val site in these cells.
Despite the
careful selection of sgRNA based on its high specificity, the results showed
low efficacy. Based on previous study, Cas9-mediated cleavage can be nullified
by single mismatches occurring at the interface between sgRNA and the target
site. This effect is predominant within the final 10-12 nucleotides situated at
the 3’ end of the 20-nucleotide sgRNA targeting region (Zhang
et al., 2017). The decrease in mRNA expression following
CRISPR/Cas9 gene editing is attributed to various factors, including
alterations in transcriptional regulation, mRNA stability, or degradation sites
(Bishop and Hawley, 2022; Javaid and Choi, 2021). The altered codons might
not have been translated into different amino acids or influenced the SOD2
conformation, causing challenges in its detection in a western blot assay.
Figure 2 Design of sgRNA targeting the Ala16Val SNP region within exon 2 of the SOD2 gene. (A) Selected sodex2.1 sgRNA sequence from exon 2 (blue box), which contains Ala16Val site in nt 401-403 (underlined codon); PAM of the target, bold fonts. (B) Cleavage activity assay in HEK293T cells showed moderate green fluorescence intensity. (C) Reduction in relative mRNA expression, while the protein expression level remained unaffected, showing no significant impact, ***p<0.001.
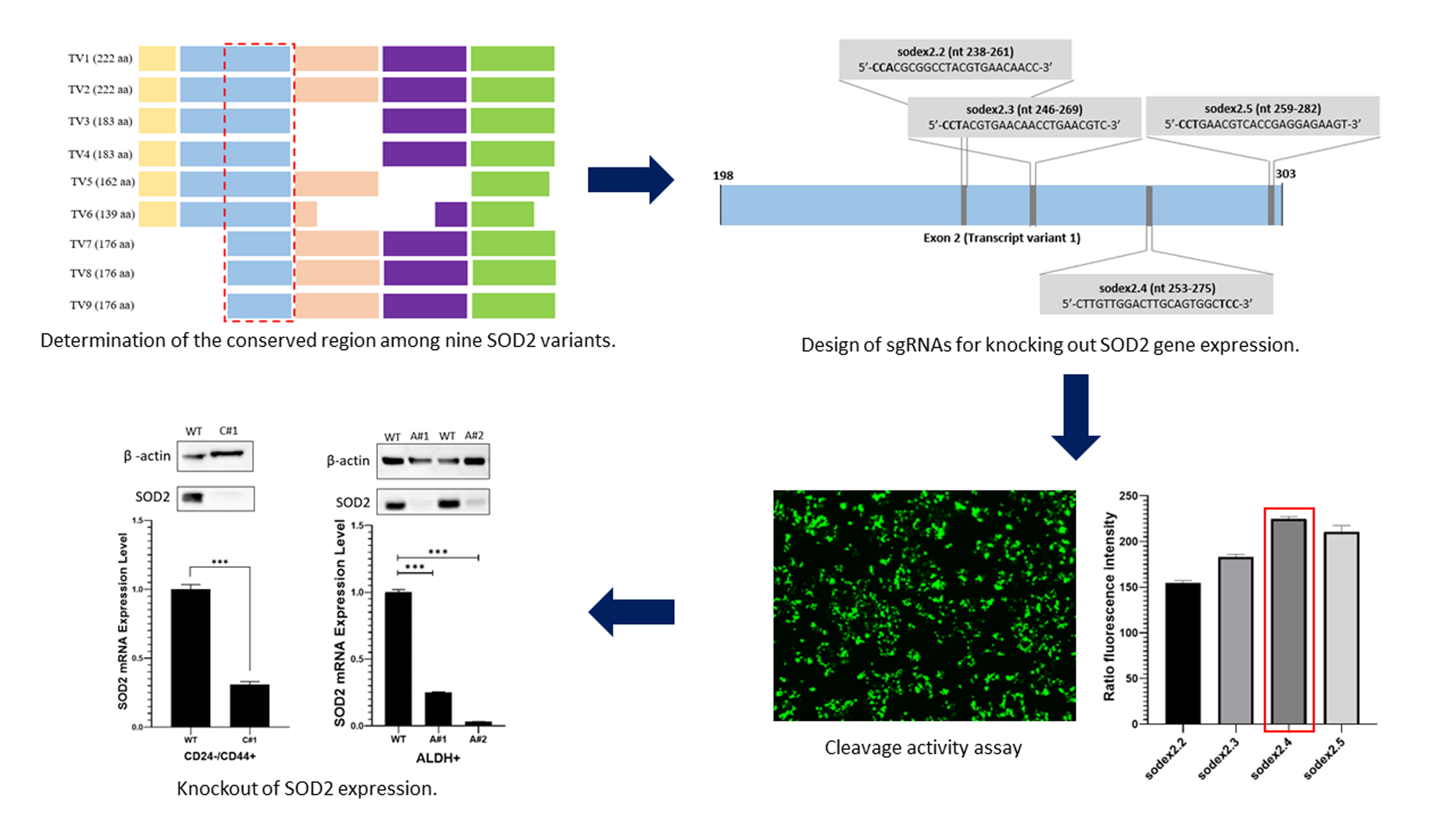
Figure 3 Position of sgRNAs targeting the conserved
region of SOD2 variants within exon 2. Four sgRNAs were selected based on the
efficiency score (max. efficiency = 1). sgRNAs sequences, grey boxes; exons,
blue box; PAM of the target, bold fonts
To
ensure the effectiveness of CRISPR/Cas9 system across all SOD2 variants, sgRNAs
targeting the conserved region among nine SOD2 variants as identified in the
NCBI database, were designed as presented in Figure Supplementary. A total of
four sgRNA candidates were obtained using CRISPRdirect online tools, described
in Figure 3, providing information about their respective on-target sites, as
shown in Table 1. According to a previous study, a greater number of on-target
sites showed a higher potential for off-target effects (Naito
et al., 2014). The characteristics of sgRNAs, including %GC
content, sgRNA length, and melting temperature also play crucial roles in
determining on-target efficacy. Previous studies have shown that sgRNAs with a
GC content ranging from 40 to 60% are effective in enhancing gene editing
efficiency using CRISPR/Cas9 system. Specifically, sgRNAs with approximately
50% GC content are recommended due to their efficiency in CRISPR gene editing (Mohammadhassan et al., 2022). Lv et al. (2019) also showed that a
20-nucleotide segment of sgRNA can be highly effective in reducing off-target
effects. However, when sgRNA is shorter than 15 nucleotides, the Cas
endonuclease may not show any activity for knocking out the target gene (Lv et al., 2019). In this study, all
sgRNAs were approximately 20 nucleotides in length, consisting of only one
target site within the 20mer+PAM, which showed a high level of specificity.
Table 1
Characteristics of four sgRNA candidates
|
sgRNA name |
Position |
%GC |
Tm (°C) |
Number of target sites | |||
|
start |
end |
20mer+PAM |
12mer+PAM |
8mer+PAM | |||
|
sodex2.2 |
517 |
539 |
65 |
78.95 |
1 |
1 |
1285 |
|
sodex2.3 |
525 |
547 |
50 |
70.89 |
1 |
3 |
789 |
|
sodex2.4 |
532 |
554 |
55 |
72.29 |
1 |
2 |
429 |
|
sodex2.5 |
538 |
560 |
55 |
73.54 |
1 |
2 |
734 |
Computational tools offer valuable
insights into sgRNA specificity. However, verifying their specificity and
efficiency through cleavage activity assay is essential. Among the four sgRNAs
tested, sodex2.4 showed the highest cleavage activity on the pEGxxFP expression
plasmid. This is evident by the strongest green fluorescence intensity of EGFP
presented in Figures 4A and 4B. This observation corresponds with the
assessment conducted using CRISPRdirect tool, which identified sodex2.4 as the
optimal sgRNA based on the number of on-target sites and GC content.
Consequently, the effectiveness of sodex2.4 sgRNA in suppressing SOD2
expression in HEK293T cells was validated. The results showed a complete
disruption of SOD2 mRNA expression to 0.3-fold and approximately complete loss
of its protein expression. This showed knockout effect of SOD2 expression
following CRISPR/Cas9 genome editing, illustrated in Figures 4C and 4D.
Further
assessment was conducted to evaluate the optimal potential of sodex2.4 sgRNA to
knockout SOD2 expression in CRISPR/Cas9 genome editing experiments conducted on
human BCSCs, specifically ALDH1+ and CD24-/CD44+ cells. The results showed a
significant reduction in SOD2 mRNA expression in CD24-/CD44+ (C#1) and ALDH1+
(A#1 and A#2) clones reaching levels of 0.3, 0.25, and 0.05, respectively,
compared to wild-type cells, as presented in Figure 5A. These results were
corroborated by protein expression analysis through Western Blot assays shown
in Figure 5B. As anticipated, all KO-SOD2 clones in CD24-/CD44+ and ALDH1+
cells showed values ranging from very low to negligible expression of SOD2
protein. This showed the successful knockout of SOD2 expression at both mRNA
and protein levels in human BCSCs.
Figure 4 The
efficiency of sgRNAs targeting the conserved region of all SOD2
variants for CRISPR/Cas9 system in HEK293T cells. (A-B) Cleavage
activity assay. (A) Fluorescent
microscope images were contrasted with cells under
a bright field. (B) Fluorescence intensity ratio of
HEK293T cells edited with sodex2.2, sodex2.3, sodex2.4, and sodex2.5
CRISPR/Cas9 system to wild-type HEK293T cells. Fluorescence intensity was
determined using ImageJ software and data obtained were represented as mean ± SD. (C) SOD2 mRNA relative expression levels in
HEK293T cells edited with sodex2.4 CRISPR/Cas9
system normalized to wild-type (WT) HEK293T.
*p<0.01. (D) SOD2 protein expression in HEK293T cells
edited with sodex2.4 CRISPR/Cas9 system compared to WT HEK293T and
Figure 5 Effect of CRISPR/Cas9 genome editing of SOD2 in human CD24-/CD44+ and ALDH1+
BCSCs. (A)
mRNA expression levels of SOD2 in BCSCs were analyzed using qRT-PCR. (B) Protein expression of
SOD2 in BCSCs analyzed using Western Blot assay.
WT stands for wild-type, C#1, A#1, and A#2 were SOD2 knockout
(SOD2-KO) clones in the respective BCSCS. actin was used as a housekeeping protein. All data of SOD2-KO were
compared to the wild-type cells, without genome editing,
***p<0.001.
CRISPR/Cas9 system is
widely acclaimed in the field of genetic engineering for its precision,
effectiveness, and affordability in genome editing. However, design of specific
and efficient sgRNA remains a challenging endeavor, requiring careful
consideration of gene variations. In conclusion, we successfully designed four
sgRNAs to knockout the SOD2 gene expression in BCSCs using CRISPR/Cas9
technology. Among these four sgRNAs, we chose sgRNA sodex2.4, which has the
highest efficiency of cleavage activity, to verify the knockout effect at the
mRNA and protein levels. These findings highlight that precise sgRNA design is
important to specifically target the CRISPR/Cas9 genome editing. Further
studies are needed to elaborate the impact of KO SOD2 on the aggressiveness of
BCSCs.
This study received
support from the PUTI Q2 Batch 2 Research Grant (Number: KB-1252/UN2.RST/HKP.05.00/2022)
provided by the Directorate for Research and Community Engagement at
Universitas Indonesia.
| Filename | Description |
|---|---|
| R2-CE-6680-20240131150129.docx | --- |
Abdelrauf, L.M., Rahman, M.F.A., Abdel-Maksoud
S.M., Farag N.M., Hashad I.M., 2017. Association of Manganese Superoxide
Dismutase Ala16Val Polymorphism in The Incidence of Acute Myocardial Infarction
in The Egyptians. Journal of Genetic Engineering and Biotechnology, Volume
15(2), pp. 415–418
Ahmad, Y., Zain, Z., Aziz, N., 2018. Multistage
Logistic Regression Model for Analyzing Survival From Colorectal Cancer. International
Journal of Technology, Volume 9(8), pp. 1618–1627
Amalia, R., Abdelaziz, M., Puteri, M.U., Hwang,
J., Anwar, F., Watanabe, Y., Kato, M., 2019. TMEPAI/PMEPA1 Inhibits Wnt
Signaling by Regulating Beta-Catenin Stability and Nuclear Accumulation in
Triple Negative Breast Cancer Cells. Cellular Signalling, Volume 59, pp. 24–33
Aryal, N.K., Wasylishen, A.R., Lozano, G., 2018.
CRISPR/Cas9 Can Mediate High-Efficiency Off-Target Mutations in Mice In Vivo. Cell
Death and Disease, Volume 9(11), p. 1099
Atkins, A., Chung, C.H., Allen, A.G., Dampier,
W., Gurrola, T.E., Sariyer, I.K., Nonnemacher, M.R., Wigdahl, B., 2021.
Off-Target Analysis in Gene Editing and Applications for Clinical Translation
of CRISPR/Cas9 in HIV-1 Therapy. Frontiers in Genome Editing, Volume 3, p. 673022
Ayob, A.Z., Ramasamy, T.S., 2018. Cancer Stem
Cells as Key Drivers of Tumour Progression. Journal of Biomedical Science, Volume
25, pp. 1–18
Azadmanesh, J., Trickel, S.R., Borgstahl, G.E.O.
2017. Substrate-Analog Binding and Electrostatic Surfaces of Human Manganese
Superoxide Dismutase. Journal of Structural Biology, Volume 199(1), pp. 68–75
Bishop, D.J., Hawley, J.A., 2022. Reassessing The
Relationship Between mRNA Levels and Protein Abundance in Exercised Skeletal
Muscles. Nature Reviews Molecular Cell Biology, Volume 23(12), pp. 773–774
Cramer-Morales, K., Heer, C.D., Mapuskar, K.A., Domann,
F.E., 2015. SOD2 Targeted Gene Editing By CRISPR/Cas9 Yields Human Cells Devoid
of MnSOD. Free Radical Biology and Medicine, Volume 89, pp. 379–386
Da-Cruz-Jung, I.E., Da-Cruz, I.B.M., Barbisan,
F., Trott, A., Houenou, L.J., Osmarin-Turra, B., Duarte, T., De Souza-Praia,
R., Maia-Ribeiro, E.A., Da-Costa-Escobar-Piccoli, J., Bica, C. G., Duarte, M.,
2020. Superoxide Imbalance Triggered by Val16Ala-SOD2 Polymorphism Increases
The Risk of Depression and Self-Reported Psychological Stress in Free-Living
Elderly People. Molecular Genetics and Genomic Medicine, Volume 8, p. e1080
Irsyad, M., Whulanza, Y., Katili, P.A.,
Antarianto, R.D., Jasirwan, C.O.M., Bugtai, N., 2022. Development of
Auto-Pivot: Automated Platform In Vitro for Cell Tissue Culture. International
Journal of Technology, Volume 13(8), pp. 1651–1662
Javaid, N., Choi, S., 2021. CRISPR/Cas System And
Factors Affecting Its Precision and Efficiency. Frontiers in Cell and
Developmental Biology, Volume 9, p. 761709
Karlapudi, A.P., Venkateswarulu, T.C.,
Tammineedi, J., Srirama, K., Kanumuri, L., Kodali, V.P., 2018. In SilicosgRNA
Tool Design for CRISPR Control of Quorum Sensing in Acinetobacter Species. Genes
and Diseases, Volume 5, pp. 123–129
Kim, Y.S., Gupta-Vallur, P., Phaëton, R.,
Mythreye, K., Hempel, N.A.-O., 2017. Insights Into The Dichotomous Regulation
of SOD2 in Cancer. Antioxidants, Volume 6(4), pp. 86–97
Konstantakos, V.A.-O., Nentidis, A., Krithara, A.,
Paliouras, G., 2022. CRISPR-Cas9 gRNA Efficiency Prediction: An Overview of
Predictive Tools and The Role of Deep Learning. Nucleic Acids Research, Volume
50, pp. 3616–3637
Livak, K.J., Schmittgen, T.D., 2001. Analysis of
Relative Gene Expression Data Using Real-Time Quantitative PCR and The Method. Methods, Volume 25, pp. 402–408
Lv, J., Wu, S., Wei, R., Li, Y., Jin, J., Mu, Y.,
Zhang, Y., Kong, Q., Weng, X., Liu, Z., 2019. The Length of Guide RNA and
Target DNA Heteroduplex Effects on CRISPR/Cas9 Mediated Genome Editing
Efficiency in Porcine Cells. Journal of Veterinary Science, Volume
20(3), p. 1125082
Malik, A., Gul, A., Munir, F., Amir, R., Alipour,
H., Babar, M.M., Bakhtiar, S.M., Paracha, R.Z., Khalid, Z., Hayat, M.Q., 2021.
Evaluating The Cleavage Efficacy of CRISPR-Cas9 sgRNAs Targeting Ineffective
Regions of Arabidopsis thaliana Genome. Peerj, Volume 9, p. E11409
Modrzejewski, D., Hartung, F., Lehnert, H.,
Sprink, T., Kohl, C., Keilwagen, J., Wilhelm, R., 2020. Which Factors Affect
The Occurrence of Off-Target Effects Caused by The Use of CRISPR/Cas: A
Systematic Review in Plants. Frontiers in Plant Science, Volume 11, p. 574959
Mohammadhassan, R., Tutunchi, S., Nasehi, N.,
Goudarziasl, F., Mahya, L., 2022. The Prominent Characteristics of the
Effective sgRNA for a Precise CRISPR Genome Editing. In: CRISPR Technology -
Recent Advances
Naito, Y., Hino, K., Bono, H., Ui-Tei, K., 2014.
CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA With Reduced
Off-Target Sites. Bioinformatics, Volume 31, pp. 1120–1123
NCBI (National Center of Biotechnology
Information), 2023. Current Theme: SOD2 superoxide dismutase 2 (Homo sapiens
(human)). Available online at
https://www.ncbi.nlm.nih.gov/gene/6648#reference-sequences, accessed on
February 5, 2023.
Nusantara, A.C., Purwanti, E., Soelistiono, S.,
2016. Classification of Digital Mammogram Based on Nearest-Neighbor Method for
Breast Cancer Detection. International Journal of Technology, Volume 7, pp.
71–77
Sari, M.I., Daulay, M., Wahyuni, D.D., 2019.
Superoxide Dismutase Levels and Polymorphism (Ala16Val) in Tuberculosis
Patients With Diabetes Mellitus in Medan City. Open Access Macedonian
Journal of Medical Sciences, Volume 7(5), p. 730
Sarkar, P., Basu, K., Sarkar, P., Chatterjee, U.,
Mukhopadhyay, M., Choudhuri, M.K., Srakar, D.K., 2018. Correlations Of Aldehyde
Dehydrogenase-1 (ALDH1) Expression with Traditional Prognostic Parameters and
Different Molecular Subtypes of Breast Carcinoma. Clujul Medical, Volume
91, pp. 181–187
Shiraishi, A., Tachi, K., Essid, N., Tsuboi, I.,
Nagano, M., Kato, T., Yamashita, T., Bando, H., Hara, H., Ohneda, O., 2017.
Hypoxia Promotes The Phenotypic Change of Aldehyde Dehydrogenase Activity of
Breast Cancer Stem Cells. Cancer Science, Volume 108(3), pp. 362–372
Srivastava, N., Usmani, S.S., Subbarayan, R.,
Saini, R., Pandey, P.K., 2023. Hypoxia: Syndicating Triple Negative Breast
Cancer Against Various Therapeutic Regimens. Frontiers in Oncology, Volume
13, p. 1199105
Wanandi, S.I., Syahrani, R.A., Arumsari, S., Wideani,
G., Hardiany, N.S., 2019. Profiling of Gene Expression Associated with Stemness
and Aggressiveness of ALDH1A1-Expressing Human Breast Cancer Cells. The
Malaysian Journal of Medical Sciences, Volume 26(5), pp. 38–52
Wanandi, S.I., Yustisia, I., Neolaka, G.M.G., Jusman,
S.W.A., 2017. Impact of Extracellular Alkalinization on The Survival of Human
CD24-/CD44+ Breast Cancer Stem Cells Associated with Cellular Metabolic Shifts.
Brazilian Journal of Medical and Biological Research, Volume 50, p. e6538
Wang, P., Zhu, Y., Xi, S., Li, S., Zhang, Y.,
2018. Association Between MnSOD Val16Ala Polymorphism and Cancer Risk: Evidence
From 33,098 Cases and 37,831 Controls. Disease Markers, Volume 2018, p. 3061974
Wardhani, B.W., Puteri, M.U., Watanabe, Y.,
Louisa, M., Setiabudy, R., Kato, M., 2020. TGF-Beta-Induced TMEPAI Attenuates
The Response of Triple-Negative Breast Cancer Cells to Doxorubicin and
Paclitaxel. Journal of Experimental Pharmacology, Volume 12, pp. 17–26
Zhang, D., Zhang, H., Li, T., Chen, K., Qiu, J.L.,
Gao, C., 2017. Perfectly Matched 20-Nucleotide Guide RNA Sequences Enable
Robust Genome Editing Using High-Fidelity Spcas9 Nucleases. Genome Biology, 18(1),
pp. 1–7
