Recombinant YebF-Cas9 Fusion Enzyme from Thermophilic Geobacillus kaustophilus Interaction with sgRNA by In Silico Method
Corresponding email: lischer.kenny@ui.ac.id
Published at : 25 Mar 2025
Volume : IJtech
Vol 16, No 2 (2025)
DOI : https://doi.org/10.14716/ijtech.v16i2.7372
Muthi'ah, H, Laksmi, FA, Nugraha, Y, Khayrani, AC, Nurhayati, RW, Ali, MSM & Lischer, K 2025, ‘Recombinant YebF-Cas9 fusion enzyme from thermophilic Geobacillus kaustophilus interaction with sgRNA by in silico method’, International Journal of Technology, vol. 16, no. 2, pp. 662-671
| Hajar Muthi`ah | Bioprocess Engineering, Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia |
| Fina Amreta Laksmi | Research Center for Applied Microbiology, National Research and Innovation Agency, Cibinong, West Java, 16915, Indonesia |
| Yudhi Nugraha | 1. Research Center for Molecular Biology Eijkman BRIN, National Research and Innovation Agency, Cibinong, West Java, 16915, Indonesia. 2. Department of Molecular Medicine, University of Pavia, 27100, |
| Apriliana Cahya Khayrani | 1. Chemical Engineering, Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia |
| Retno Wahyu Nurhayati | 1. Research Centre of Biomedical Engineering, Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia. 2. Chemical Engineering, Departmen |
| Mohd Shukuri Mohamad Ali | 1. Research Center for Applied Microbiology, National Research and Innovation Agency, Cibinong, West Java, 16915, Indonesia 2. Enzyme and Microbial Technology Research Center, Universiti Putra Mala |
| Kenny Lischer | 1. Bioprocess Engineering, Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia, Depok, West Java, 16424, Indonesia 2. Research Centre of Biomedical Engineering, Departme |
Genetic engineering is a process that changes the structure of an organism by removing, inserting, or modifying its genetic material. Currently, the most widely used method in genetic engineering is CRISPR-Cas9, representing “Clustered Regularly Interspaced Short Palindromic Repeat-Associated Protein 9“. As an intracellular enzyme, the production of Cas9 is complex and costly due to the need for extraction and purification. In comparison, YebF is a protein that can be localized extracellularly. By fusing YebF with Cas9 (YebF-Cas9), it is possible to express and localize Cas9 extracellularly. This fusion potentially alters Cas9 ability to bind with sgRNA (single guide RNA). Therefore, this study aimed to explore the interaction between sgRNA and Cas9 from Geobacillus kaustophilus fused with YebF using in silico methods. In the in silico experiment, the molecular docking method was used to determine biomolecular interactions with variations in sgRNA, namely spacer 10, 20, 30 nt, repeat 16, 25, 36 nt, and tracrRNA 63, 98, 140 nt. The results showed that changes in the length of the spacer, repeat, and tracrRNA could affect the level of binding affinity formed in YebF-Cas9-sgRNA complex from Geobacillus kaustophilus. The optimal length of the molecular docking results in terms of affinity and position was in the variation of 30 nt spacer with 16 nt repeat and 98 nt tracrRNA, with the binding affinity of –419.24 kcal/mol.
Fusion enzyme; Gerbille’s kaustophilus; Molecular docking; sgRNA; YebF-Cas9
Genetic engineering is a process that changes the structure of an organism by removing, inserting, or modifying the genetic material contained in the target object. One of the genetic engineering methods is gene editing, which enables specific modifications to an organism’s DNA sequence and allows adaptation to its genetic makeup (Fridovich-Keil, 2023; Maeder and Gersbach, 2016). The foundation of gene editing method is the hypothesis that targeted double-stranded breaks (DSBs) in DNA can stimulate endogenous cellular repair pathways, allowing exploitation to introduce specific mutations or precise edits to the genome (Doudna, 2020; Kosicki et al., 2018). The Clustered Regularly Interspaced Short Palindromic Repeat-Associated Protein 9 (CRISPR-Cas9) method is considered the simplest and most efficient, providing several advantages compared to previous methods. The advantages include simple design, high level of target specificity, reduced off-target toxins, ability to target several genes, and ease of delivery to cells (Ahmad et al., 2022; Doudna and Charpentier, 2014; Feng et al., 2013). The CRISPR-Cas9 method is a combination of the Cas9 enzyme with a piece of RNA called sgRNA (single guide-Ribonucleid Acid) (Barrangou and Doudna, 2016). Cas9 acts as a restriction enzyme or endonuclease that cuts DNA at certain positions according to the combination of sgRNA (Doudna and Charpentier, 2014). Several foreign DNA sequences can be integrated into the CRISPR locus and transcribed into CRISPR RNA (crRNA) (Koonin and Makarova, 2019). Subsequently, the crRNA will combine with the trans-activating crRNA (tracrRNA) to specifically cut the foreign DNA sequence (Koonin and Makarova, 2019). This system has been simplified for genetic engineering applications, and now includes only Cas9 nuclease and a single guide RNA (sgRNA) containing crRNA and tracrRNA elements. (Gaj et al., 2016).
Production of the Cas9 enzyme is carried out intracellularly by inserting the Cas9 gene into a plasmid which is generated by the host cell (Arumsari et al., 2024; Jinek et al., 2012). This method is often used because of its ease in genetic manipulation and high stability of expression, although there are several drawbacks. Production of intracellular Cas9 requires a more complicated process and higher costs, including complex purification method (Doudna and Charpentier, 2014). This is due to the need to separate the Cas9 enzyme from other cellular components, which increases the cost and risk of contamination (Hsu et al., 2014). Therefore, obtaining a new production method is important for commercial and clinical applications due to the efficiency and lower costs as well as high adaptability, making it more suitable for industrial-scale production (Huang et al., 2023).
One promising method is the use of Geobacillus kaustophilus, a gram-positive thermophilic bacterium that can be found in the Ring of Fire area (Ring of Fire) Pacific, including Indonesia. This bacterium grows at higher temperatures from 48 to 74°C (optimal at 60°C) and is considered a prospective chassis for establishing high-temperature resistant cell factories (Mori et al., 2022; Lischer et al., 2020a). In previous studies, Geobacillus kaustophilus was found in the Cisolong hot springs, Banten, containing Cas9 enzyme (Angela, 2022). It also was studied for the length of sgRNA suitable for Cas9 by using the molecular docking method (Pramayuditya, 2023).
Cas9 protein is known as intracellular enzyme (Qiao et al. 2019). There is a significant drawback regarding the production of intracellular enzyme such as Cas9, particularly economic aspect during industrial processes (Rosazza et al., 2023; Ferrari et al., 2023). This is due to additional unit operation for the extraction centrifugation process (Junker et al., 2001). In this context, several methods are been used to express Cas9 extracellularly and eliminate unit operation which directly affects lower capital expenditure (Ferrari et al., 2023; Junker et al., 2001). Therefore, recombinant Cas9 enzyme was fused with YebF carrier in this study. YebF is known as protein that can be localized into extracellular (Dai et al., 2021; Zhang et al., 2006). By fusing YebF with Cas9 (YebF-Cas9), it can be expressed and localized to extracellular space, simplifying the production process.
The fusion of YebF-Cas9 can affect its interaction with sgRNA, showing the need for further investigation. Due to the lack of previous related reports, the molecular docking method was used with variations of sgRNA. The results observed were analyzed in the form of the structure of YebF-Cas9-sgRNA Geobacillus kautophilus complex. According to (Husnawati et al., 2023, Sahlan et al., 2020, Lischer et al., 2020b), the majority of molecular docking studies were typically conducted to observe the interactions between drugs and their biological targets such as allosteric modulators and drug-binding affinities. However, this study applies a different method by focusing on the interaction between sgRNA and the Cas9 as well as dCas9 enzyme in silico. The results provide insights into the binding affinity and structural positioning of these molecular complexes. After identifying the suitable sgRNA, it offers new genetic tools in genetic engineering for multiple organisms.
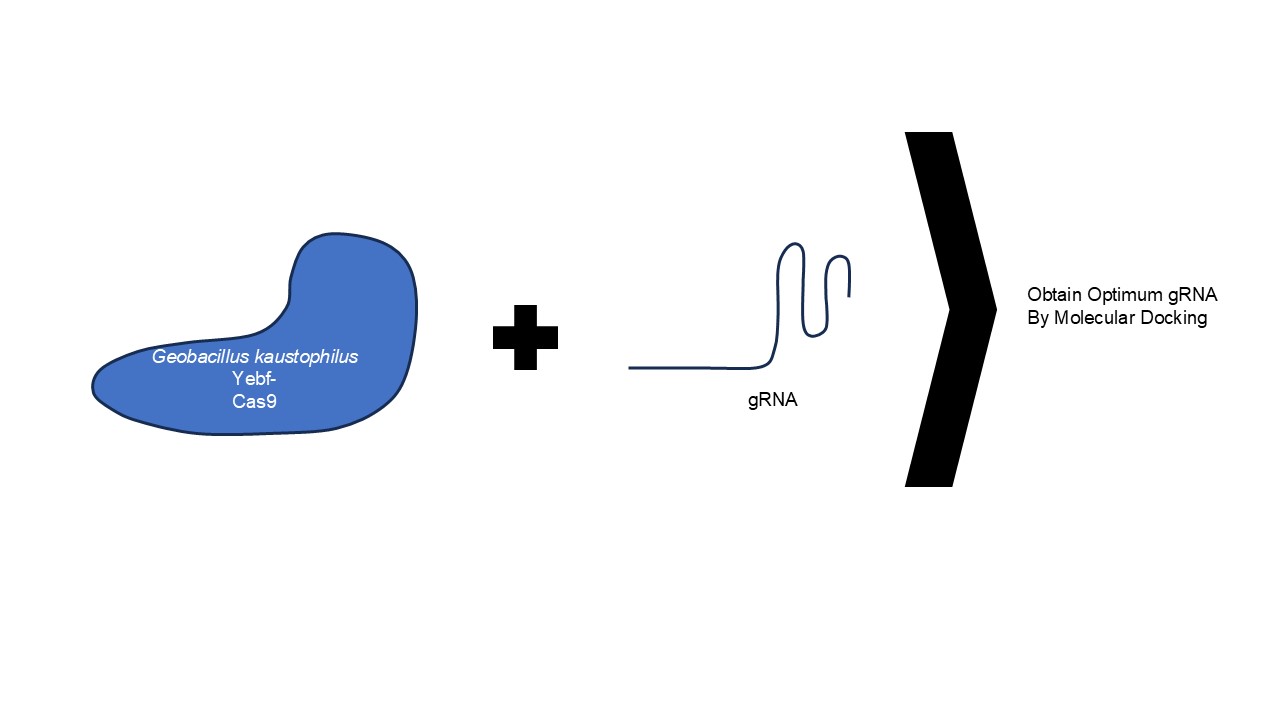
The experiment is divided into three major tasks, where two are conducted in parallel, namely preparation of YebF-Cas9 fusion enzyme sequence of Geobacillus kaustophilus and sgRNA variation. This is followed by 3D visualization and molecular docking simulation for each variant with respective variations (Figure 1).
2.1. Preparation of YebF-Cas9 Fusion Enzyme Sequence of Geobacillus kaustophilus and sgRNA Variation
The Cas9 sequence from Geobacillus kaustophilus used in this study was obtained with a length of 865 aa (amino acids) (Angela, 2022). Meanwhile, YebF enzyme sequence was obtained from ncbi at https://www.ncbi.nlm.nih.gov/gene/946363. The sgRNA sequence variations, including spacer, repeat, and tracrRNA, were sourced from previous studies (Pramayuditya, 2023). The data obtained were used to examine the biomolecular interactions occurring with YebF-Cas9 enzyme Geobacillus kaustophilus.
2.2. Visualization of 3D Structure of YebF-Cas9 Fusion Enzyme of Geobacillus kaustophilus
The 3D structure of YebF-Cas9 fusion enzyme from Geobacillus kaustophilus was generated using the I-TASSER platform (accessed at: https://zhanggroup.org/I-TASSER/) (Zhou et al., 2022; Zheng et al., 2021; Yang and Zhang, 2015). The FASTA format file containing YebF-Cas9 sequence is uploaded by pressing the “choose file” button. After the data is successfully uploaded, the user clicks “Run I-TASSER”. The data processing process continues until the site states that the 3D structure can be downloaded.
Visualization of 2D and 3D Structure of sgRNA Variations
The visualization of sgRNA structure differs from YebF-Cas9, which includes generating the secondary (2D) structure, followed by the tertiary (3D) structure. To create a sequential structure, all sgRNA variations are inputted into the RNAfold website (accessed at http://www.unafold.org/mfold/applications/rna-folding-form.php) to form sgRNA fold structure, namely in the repeat and tracrRNA sections (Zuker, 2023). The sequence file is entered into the website, then press "proceed". After a few minutes, the website provides a sequential structure image along with Vienna data in the form of a .b file. The contents of the Vienna data in the form of a .b file are a collection of points of a number of nucleotides in the sequence. Subsequently, these two outputs are saved to be used for the tertiary structure creation stage. To create a tertiary structure, the RNAcomposser website is used (accessed at https://rnacomposer.cs.put.poznan.pl/) and input the Vienna data (Sarzynska et al., 2023; Popenda et al., 2012). The sequence is inputted manually then press "compose". The results are obtained after waiting 1-5 minutes and the tertiary structure image data along with .pdb data will be obtained.
Molecular Docking Simulations
The molecular docking process was performed using HDOCK (Yan et al., 2020), following these steps: The 3D structure pdb data of YebF-Cas9 from Geobacillus kaustophilus were submitted as the receptor in “Input Receptor Molecule”. The 3D structure pdb data of sgRNA were also submitted as the ligand in “Input Ligand Molecule”. The interaction site residues of sgRNA within Cas9 were manually entered through “Advanced Options (Optional)”> Receptor Binding Site Residue(s) section. The residues were entered in the format M:A, where “M” represented the residue number of the protein interaction site and “A” was the protein chain. The interaction sites followed the procedures by Sun et al., (2019). A name was given to the file, and a personal email address was provided for docking results to be sent directly to the specified address after completion. The process was initiated by clicking submit to start the molecular docking. The results included the top 10 models in 3D, along with binding affinity values (docking score), confidence score, ligand RMSD, and interface residues. These docking results could be downloaded by clicking “All the results in a package”.
Figure 1 Scheme of docking process Yebf-Cas9 with sgRNA
Docking simulations were conducted by treating enzyme as a rigid unit and allowing sgRNA to remain flexible. Moreover, flexibility of sgRNA molecule is very important to achieve high binding affinity at the appropriate position. This is explained by Sun et al., (2019) namely, the spacer must enter the HNH and RuvC areas. Each docking result carried out on the HDOCK server produces the 10 best options for interaction modeling. However, only the best of each variation will be discussed. The position of sgRNA against YebF-Cas9 from Geobacillus kaustophilus and its binding affinity from modeling will be the main indicators in selecting the best model. Visually, the 3D .pdb file of YebF-Cas9-sgRNA complex from docking can be viewed using PyMOL or directly through the web server. The coloring guide can be found in the legend of Table 1 below.
Table 1 Colouring Guide of YebF-Cas9 Geobacillus kaustophilus complex structure.
3.1. Spacer Variations
Based on docking results, the best model was obtained for each spacer variation. For spacer variations, model 3 was obtained for the 10 nt spacer, model 1 for the control (20 nt spacer), and model 1 for the 30 nt spacer. Binding affinity values for the models are listed in Table 2.
Spacer variation 10 nt (Figure 2a) is considered superior both in position and binding affinity because the tip tends to enter the HNH and RuvC areas. Meanwhile, the 20 and 30 nt (Figures 2b and 2c) are not considered superior because of their distance from the HNH and RuvC areas. However, the 30 nt spacer variation appears significantly close to the HNH and RuvC areas compared to the 20 nt. The 30 nt spacer variation also has advantages in binding affinity and confidence score values due to higher values than other spacer variations. The control variation model has the lowest level of binding affinity compared to other spacer variations. This shows that the 30 nt spacer variation is the most suitable length for sgRNA spacer in -Cas9-sgRNA complex of Geobacillus kaustophilus.
Table 2 Binding Affinity of YebF-Cas9-sgRNA Geobacillus kautophilus Docking Models with Spacer Variations.
Figure 2 Complex Structure from Docking Models of YebF-Cas9-sgRNA Geobacillus kaustopilus Spacer Variations (a) 10 nt, (b) 20 nt, (c) 30 nt (blue arrow for Yebf-Cas9 and Green arrow for sgRNA).
3.2. Repeat Variations
The best model was achieved for each repeat variation, where model 3 was obtained for the 16 nt (control) and 25 nt repeat variation, as well as model 1 for the 36 nt spacer. Binding affinity values for the models are listed in Table 3.
Table 3 Binding Affinity of YebF-Cas9-sgRNA Geobacillus kautophilus Docking Models with Repeat Variations
Repeat variations 16 nt, 25 nt, and 36 nt (Figures 3a, 3b, and 3c) appear positionally superior because their spacer ends tend to enter the area HNH and RuvC. However, when observed from the aspect of binding affinity values, the 25 nt repeat variation has the most negative value compared to others. This suggests that 25 nt repeat variation is the most suitable length for sgRNA repeat in YebF-Cas9-sgRNA Geobacillus kautophilus complex.
3.3. TracRNA Variations
Based on docking results, the best model was obtained for each tracrRNA variation. Specifically, model 1 was obtained for the 63 nt tracrRNA variation, model 3 for the control (98 nt tracrRNA), and model 2 for the 140 nt tracrRNA. Binding affinity values for the models are listed in Table 4.
The 63 nt tracrRNA variation (Figure 4a) cannot be favored by position or by binding affinity. This is because the end of the spacer only binds to the RuvC area. The 98 nt tracrRNA variation (Figure 4b) has an advantage due to the presence of a fairly negative binding affinity value and positionally the tip of the spacer can be observed entering the RuvC and HNH areas. Meanwhile, the 140 nt tracrRNA variation (Figure 4c) appears superior because it has the most negative binding affinity value compared to others. In terms of the spacer end position, 140 nt tracrRNA variation tends to enter YebF-Cas9-sgRNA complex of Geobacillus kaustophilus. However, the inclusion of the spacer end of the 140 nt tracrRNA variation did not appear to bind the HNH area. This can limit the DNA-cutting process normally induced by double-strand cuts, leading to very low or non-existent gene editing efficiency. Therefore, tracrRNA variations 98 nt is the most suitable length for tracrRNA in sgRNAs in YebF-Cas9-sgRNA complex of Geobacillus kautophilus.
Figure 3 Complex Structure from Docking Models of YebF-Cas9-sgRNA Geobacillus kaustopilus Repeat Variations (a) 16 nt, (b) 25 nt, (c) 36 nt (blue arrow for Yebf-Cas9 and Green arrow for sgRNA).
Table 4 Binding Affinity of YebF-Cas9-sgRNA Geobacillus kautophilus Docking Models with TracrRNA Variations.
Figure 4 Complex Structure from Docking Models of YebF-Cas9-sgRNA Geobacillus kaustopilus TracrRNA Variations (a) 63 nt, (b) 98 nt, (c) 140 nt (blue arrow for Yebf-Cas9 and Green arrow for sgRNA).
Discussion
The results from this study show that variations in spacer and tracrRNA lengths have a significant influence on binding affinity and position in the complex structure. Figure 5 shows the binding affinity results while Figure 6 presents the confidence score value of the molecular docking. Based on the results, the best affinity and confidence score values are for the 30 nt spacer variation. Similarly, Sun et al., (2019) stated that sgRNA flexibility is very important to achieve high binding affinity in the HNH and RuvC regions. Harrington et al., (2017) also showed that crRNA sequences formed one fold while tracrRNA could form more than one fold, affecting the stability and efficiency of sgRNA-Cas9 complex.
Figure 5 Graph of Binding Affinity (unit: kcal/mol) Molecular Docking Results for Each Variation.
Figure 6 Binding Confidence Score Graph from Molecular Docking Results for Each Variation.
Table 5 shows a comparison between the results of the optimal length of sgRNA in terms of binding affinity. In this study, a spacer length of 30 nt was proven to be optimal both in terms of binding affinity and position compared to others. Meanwhile, Pramayuditya (2023) showed that sgRNA design with variations in spacer and tracrRNA lengths could influence the number of hydrogen bonds formed in sgRNA-Cas9 complex, thereby affecting the binding affinity and stability of the complex. These results showed that appropriate spacer and tracrRNA lengths are critical for the efficiency and stability of sgRNA-Cas9 complex.
Table 5 Comparison of sgRNA Length and Affinity with Recent Studies.
In conclusion, this study showed that modifications in the lengths of the spacer, repeat, and tracrRNA significantly influenced the binding affinity in YebF-Cas9-sgRNA complex from Geobacillus kaustophilus. The optimal length of the molecular docking results in terms of affinity and position was in the variation of 30 nt spacer, 16 nt repeat, and 98 nt tracrRNA. The optimum sgRNA of YebF-Cas9 was different compared to sgRNA for Cas9 only. This study served as the first to unleash the potential of YebF-Cas9 from Geobacillus kaustophilus. Therefore, further investigations should be conducted to confirm interactions between YebF-Cas9 and optimum sgRNA by observing the ability for genetic engineering in vitro and in vivo.
The author is grateful to individuals and institutions, including the supervisors who provided guidance throughout the writing process, thesis examiners, Research Center for Applied Microbiology, National Research and Innovation Agency (BRIN), and the Department of Chemical Engineering, Faculty of Engineering, Universitas Indonesia that participated in the process. This study was supported by a research grant of national collaboration (PKDN) by Kemendibukristek, Indonesia Number: NKB-1148/UN2.RST/HKP.05.00/2023.
Author Contributions
HM draft first paper and analysis of the result. KL, FA, YN, do conceptual, analysis, supervise, and finalization draft of paper. ACK, RWN do conceptual design and analysis. MSMA works on finalization of paper.
Conflict of Interest
There is no conflict of interest.
Ahmad, A, Khan, SH & Khan, Z 2022, The CRISPR/Cas tool kit for genome editing, Springer, https://doi.org/10.1007/978-981-16-6305-5
Angela, K 2022, Identification of thermophilic bacteria isolated from Cisolong Hot Spring Banten based on lab-on-chip technology, Universitas Indonesia
Arumsari, S, Wanandi, IS, Syahrani, RA, Watanabe, Y & Mizuno, S 2024, ‘Design of specific and efficient sgRNA for CRISPR/Cas9 system to knockout superoxide dismutase 2 in breast cancer stem cells’, International Journal of Technology, vol. 15, no. 2, pp. 353-363, https://doi.org/10.14716/ijtech.v15i2.6680
Barrangou, R & Doudna, J 2016, ‘Applications of CRISPR technologies in research and beyond’, Nature Biotechnology, vol. 34, pp. 933-941, https://doi.org/10.1038/nbt.3659
Dai, A, Wu, Z, Zheng, R & Zheng, Y 2021, ‘Extracellular expression of natural cytosolic nitrilase from Rhodococcus zopfii through constructing a transmembrane tunnel structure in Escherichia coli cells’, Process Biochemistry, vol. 103, pp. 71-77, https://doi.org/10.1016/j.procbio.2021.02.003
Doudna, JA & Charpentier, E 2014, ‘The new frontier of genome engineering with CRISPR-Cas9’, Science, https://doi.org/10.1126/science.1258096
Doudna, JA 2020, ‘The promise and challenge of therapeutic genome editing’, Nature, vol. 578, pp. 229–236, https://doi.org/10.1038/s41586-020-1978-5
Feng, Z, Zhang, B, Ding, W, Liu, X, Yang, D, Wei, P, Cao, F, Zhu, S, Zhang, F, Mao, Y & Zhu, J 2013, ‘Efficient genome editing in plants using a CRISPR/Cas system’, Cell Research, vol. 23, pp. 1229–1232, https://doi.org/10.1038/cr.2013.114
Ferrari, E, Jarnagin, AS & Schmidt, BF 1993, Commercial production of extracellular enzymes, Willey, https://doi.org/10.1128/9781555818388.ch62
Fridovich-Keil, JL 2023, Gene editing, viewed 7 July 2024 (https://www.britannica.com/science/gene-editing)
Gaj, T, Sirk, SJ, Shui, S-L & Liu, J 2016, ‘Genome-editing technologies: principles and applications’, Cold Spring Harbor Perspectives in Biology, vol. 8, no. 12, https://doi.org/10.1101/cshperspect.a023754
Harrington, LB, Paez-Espino, D, Staahl, BT, Chen, JS, Ma, E, Kyrpides, NC & Doudna, JA 2017, ‘A thermostable Cas9 with increased lifetime in human plasma’, Nature Communications, vol. 8, no. 1, pp. 1–7, https://doi.org/10.1038/s41467-017-01408-4
Hsu, PD, Lander, ES & Zhang, F 2014, ‘Development and applications of CRISPR-Cas9 for genome engineering’, Cell, https://doi.org/10.1016/j.cell.2014.05.010
Huang, X, Li, A, Xu, P, Yu, Y, Li, S, Hu, L & Feng, S 2023, ‘Current and prospective strategies for advancing the targeted delivery of CRISPR/Cas system via extracellular vesicles’, Nanomedicine: Nanotechnology, Biology and Medicine, https://doi.org/10.1186/s12951-023-01952-w
Husnawati, Kusmardi, K, Kurniasih, R, Hasan, AZ, Andrianto, D, Julistiono, H, Priosoeryanto, BP, Artika, IM & Salleh, MN 2023, ‘Investigation of chemical compounds from Phomopsis extract as anti-breast cancer using LC-MS/MS analysis, molecular docking, and molecular dynamic simulations’, International Journal of Technology, vol. 14, no. 7, pp. 1476–1486, https://doi.org/10.14716/ijtech.v14i7.6696
Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA & Charpentier, E 2012, ‘A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity’, Science, vol. 337, no. 6096, pp. 816–821, https://doi.org/10.1126/science.1225829
Junker, B, Moore, J, Sturr, M, McLoughlin, K, Leporati, J, Yamazaki, S, Chartrain, M & Greasham, R 2001, ‘Pilot-scale production of intracellular and extracellular enzymes’, Bioprocess and Biosystem Engineering, vol. 24, pp. 39-49, https://doi.org/10.1007/s004490100230
Koonin, EV & Makarova, S 2019, ‘Origins and evolution of CRISPR-Cas system’, Philosophical Transactions B, vol. 374, pp. 1-16, http://dx.doi.org/10.1098/rstb.2018.0087
Kosicki, M, Tomberg, K & Bradley, A 2018, ‘Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements’, Nature Biotechnology, vol. 36, no. 8, pp. 765–771, https://doi.org/10.1038/nbt.4192
Lischer, K, Putra, ABRD, Wirawan, B, Avilla, F, Sitorus, SG, Nugraha, Y & Sarmoko 2020, ‘Short communication: The emergence and rise of indigenous thermophilic bacteria exploration from hot springs in Indonesia’, Biodiversitas, vol. 21, no. 11, pp. 5474-5481, https://doi.org/10.13057/biodiv/d211156
Lischer, K, Tansil, KP, Ginting, MJ, Sahlan, M, Wijanarko, A & Yohda, M 2020, ‘Cloning of DNA polymerase I Geobacillus thermoleovorans SGAir0734 from a Batu Kuwung hot spring in Escherichia coli’, International Journal of Technology, vol. 11, no. 5, pp. 921–930, https://doi.org/10.14716/ijtech.v11i5.4311
Maeder, ML & Gersbach, CA 2016, ‘Genome-editing technologies for gene and cell therapy’, Molecular Therapy, vol. 24, no. 3, pp. 430-446, https://doi.org/10.1038/mt.2016.10
Mori, K, Fukui, K, Amatsu, R, Ishikawa, S, Verrone, V, Wipat, A, Meijer, WJJ & Yoshida, K 2022, ‘A novel method for transforming Geobacillus kaustophilus with a chromosomal segment of Bacillus subtilis transferred via pLS20-dependent conjugation’, Microbial Cell Factories, vol. 21, no. 1, pp. 1–8, https://doi.org/10.1186/s12934-022-01759-8
Popenda, M, Szachniuk, M, Antczak, M, Purzycka, KJ, Lukasiak, P, Bartol, N, Blazewicz, J & Adamiak, RW 2012, ‘Automated 3D structure composition for large RNAs’, Nucleic Acids Research, vol. 40, no. 14, p. e112, https://doi.org/10.1093/nar/gks339
Pramayuditya, M 2023, Molecular docking of sgRNA with variety of spacer spacer repeat dan tracrRNA for CRISPR-Cas9 and dCas9 Geobacillus kaustophillus production, Universitas Indonesia
Qiao, J, Li, W, Lin, S, Sun, W, Ma, L & Liu, Y 2019, ‘Co-expression of Cas9 and single-guided RNAs in Escherichia coli streamlines production of Cas9 ribonucleoproteins’, Communication Biology, vol. 2, no. 161, pp. 1-6, https://doi.org/10.1038/s42003-019-0402-x
Rosazza, T, Eigentler, L, Earl, C, Davidson, FA & Stanley-Wall, NR 2023, ‘Bacillus subtilis extracellular protease production incurs a context dependent cost’, Molecular Microbiology, vol. 120, no. 2, pp. 105-121, https://doi.org/10.1111/mmi.15110
Sahlan, M, Al Faris, MNH, Aditama, R, Lischer, K, Khayrani, AC & Pratami, DK 2020, ‘Molecular docking of South Sulawesi propolis against fructose 1,6-bisphosphatase as a type 2 diabetes mellitus drug’, International Journal of Technology, vol. 11, no. 5, pp. 910–920, https://doi.org/10.14716/ijtech.v11i5.4332
Sahlan, M, Dewi, L, Pratami, DK, Lischer, K & Hermansyah, H 2023, ‘In silico identification of propolis compounds potential as COVID-19 drug candidates against SARS-CoV-2 spike protein’, International Journal of Technology, vol. 14, no. 2, pp. 387-390, https://doi.org/10.14716/ijtech.v14i2.5052
Sarzynska, J, Popenda, M, Antczak, M & Szachniuk, M 2023, ‘RNA tertiary structure prediction using RNAComposer in CASP15’, Proteins: Structure, Function, and Bioinformatics, vol. 91, no. 12, pp. 1790-1799, https://doi.org/10.1002/prot.26578
Sun, W, Yang, J, Cheng, Z, Amrani, N, Liu, C, Sheng, G, Yang, Y, Lou, J & Sontheimer, EJ 2019, ‘Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states’, Molecular Cell, vol. 76, no. 6, pp. 938–952, https://doi.org/10.1016/j.molcel.2019.09.025
Yan, Y, Tao, H, He, J & Huang, SY 2020, ‘The HDOCK server for integrated protein-protein docking’, Nature Protocols, vol. 15, pp. 1829-1852, https://doi.org/10.1038/s41596-020-0312-x
Yang, J & Zhang, Y 2015, ‘I-TASSER server: new development for protein structure and function predictions’, Nucleic Acids Research, vol. 43, no. 1, pp. 174-181, https://doi.org/10.1093/nar/gkv342
Zhang, G, Brokx, S & Weiner, JH 2006, ‘Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli’, Nature Biotechnology, vol. 24, pp. 100-104, https://doi.org/10.1038/nbt1174
Zheng, W, Zhang, C, Li, Y, Pearce, R, Bell, EW & Zhang, Y 2021, ‘Folding non-homology proteins by coupling deep-learning contact maps with I-TASSER assembly simulations’, Cell Reports Methods, vol. 1, no. 3, 100014, https://doi.org/10.1016/j.crmeth.2021.100014
Zhou, X, Zheng, W, Li, Y, Pearce, R, Zhang, C, Bell, EW, Zhang, G & Zhang, Y 2022, ‘I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction’, Nature Protocols, vol. 17, pp. 2326-2353, https://doi.org/10.1038/s41596-022-00728-0
Zuker, M 2003, ‘Mfold web server for nucleic acid folding and hybridization prediction’, Nucleic Acids Research, vol. 31, no. 1, pp. 3406-3415, https://doi.org/10.1093/nar/gkg595
