Exploring the Anti-Breast Cancer Potential of Chalcomoracin, Guangsangon E, and Morushalunin: A Computational Analysis of Compounds from Morus sp.
Corresponding email: rani.wardani1708@ui.ac.id
Published at : 07 Dec 2023
Volume : IJtech
Vol 14, No 7 (2023)
DOI : https://doi.org/10.14716/ijtech.v14i7.6707
Hakim, R.W., Putri, R.C., Fachri, W., Fadilah, F., Krisnamurti, D.G.B., Fitriani, R., Hakim, E.H., Wulansari, D., 2023. Exploring the Anti-Breast Cancer Potential of Chalcomoracin, Guangsangon E, and Morushalunin: A Computational Analysis of Compounds from Morus sp. International Journal of Technology. Volume 14(7), pp. 1586-1595
| Rani Wardani Hakim | 1. Department of Medical Pharmacy, Faculty of Medicine, Universitas Indonesia, DKI Jakarta, 10430, Indonesia, 2. Drug Discovery Research Cluster, IMERI, Faculty of Medicine, Universitas Indonesia, DKI |
| Rizky Clarinta Putri | 1. Department of Medical Pharmacy, Faculty of Medicine, Universitas Indonesia, DKI Jakarta, 10430, Indonesia, 2. Master’s Program in Biomedical Sciences, Faculty of Medicine, Universitas Indonesia, DK |
| Wilzar Fachri | 1. Department of Medical Pharmacy, Faculty of Medicine, Universitas Indonesia, DKI Jakarta, 10430, Indonesia, 2. Drug Discovery Research Cluster, IMERI, Faculty of Medicine, Universitas Indonesia, DKI |
| Fadilah Fadilah | Department of Medical Chemistry, Faculty of Medicine Universitas Indonesia, DKI Jakarta, 10430 |
| Desak Gede Budi Krisnamurti | 1. Department of Medical Pharmacy, Faculty of Medicine, Universitas Indonesia, DKI Jakarta, 10430, Indonesia, 2. Drug Discovery Research Cluster, IMERI, Faculty of Medicine, Universitas Indonesia, DKI |
| Rizki Fitriani | Organic Chemistry Division, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung, Natural Product Research Group , Jalan Ganesha 10, Bandung, 40132, Indonesia |
| Euis Holisotan Hakim | Organic Chemistry Division, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung, Natural Product Research Group , Jalan Ganesha 10, Bandung, 40132, Indonesia |
| Dewi Wulansari | Graduation School of Medicine, University of Tokyo, 1404-1 Katakuramatchi, Tokyo, Japan |
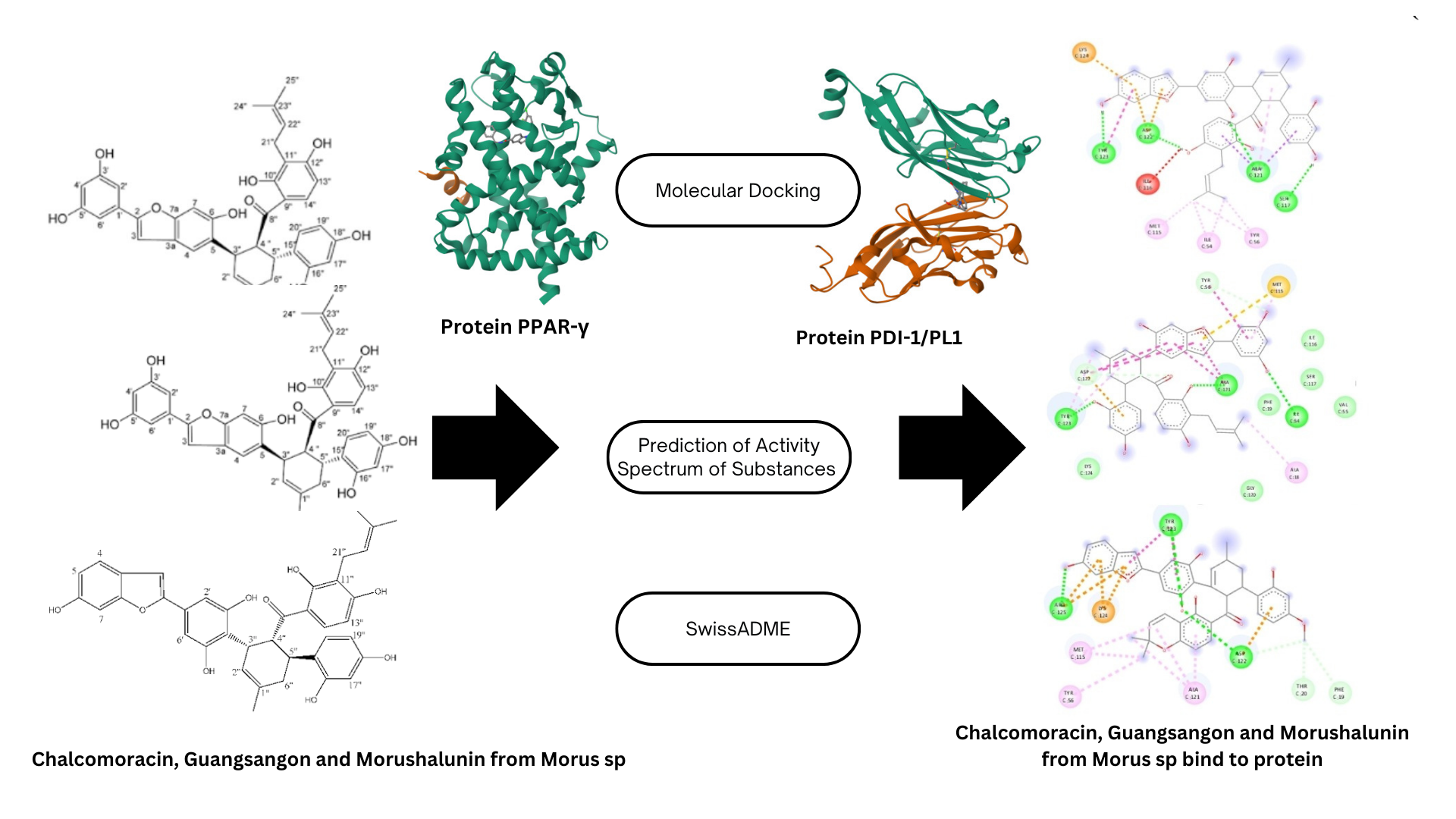
Morus sp is a plant containing
polyphenol compounds such as Chalcomoracin, Morushalunin, and Guangsangon E.
These compounds play a crucial role in modifying proteins and signaling
pathways that influence the progression of cancer cells, including breast cancer.
Therefore, this study aimed to analyze the interaction between Chalcomoracin,
Morushalunin, and Guangsangon E on PD-1 and PPAR proteins as well as
determine the physicochemical and pharmacological properties of these
compounds. To achieve this, molecular docking was conducted on PD-1 (PDB ID:
57w9) and PPAR
(PDB ID: 5two) human proteins. The results showed that
Chalcomoracin and Guangsangon E had binding capabilities to both PD-1 and
PPAR
, while Morushalunin interacted exclusively with PD-1 protein. The
interaction between Guangsangon E and PPAR
was -12.29 (Kcal/mol),
and for Chalcomoracin with PPAR
, it was -5.69 (Kcal/mol). Docking scores for
Chalcomoracin, Morushalunin, and Guangsangon E on PD-1 were -6.21 kcal/mol,
-8.91 kcal/mol, and -9.28/kcal/mol, respectively. Based on PASS analysis,
Morushalunin had potential as an HIF1a-inhibitor, while Chalcomoracin
demonstrated activity as an MMP-9 expression inhibitor. Guangsangon E showed
activity on both proteins. Additionally, drug-likeness score (DLS) for
Chalcomoracin, Morushalunin, and Guangsangon E were 1.14, 1.09, and 0.79,
respectively. These concluded that the compounds could effectively interact
with PD-1 and PPAR
two important proteins in breast cancer.
Breast cancer; Morus sp; PD-1/PDL-1 ; PPAR- ; Triple negative breast cancer
According to Globocan data from (2020), the incidence of new breast cancer cases in Indonesia reached
68.858 out of 396.914, accounting for 16.6% (Sung et al., 2021; Giaquinto et al., 2022). Peroxisome
proliferator-activated receptor gamma (PPAR), a
component of the nuclear receptor superfamily,
functions
as a transcription factor and is implicated in cancers. Additionally, PD-ligand 1 or Programmed cell death protein 1
(PD-1)
Figure 1 Chemical Structures of (a) Chalcomoracin,
(b) Guangsangon E, (c) Morushalunin from Morus sp
A study showed that the inhibitory effects of the
three compounds on leukemia P-388 cells, with Morushalunin, Guangsangon E, and
Chalcomoracin having IC50 values of 0.7 ppm, 2.5 ppm, and 1.7 ppm,
respectively (Fitriani,
Happyana and Hakim, 2021). Chalcomoracin
demonstrated the ability to inhibit the MDA-MB-231 breast cancer cell line with
an IC50 of 6 µM. However, investigations on the anticancer potential
of these compounds remain limited. Molecular docking, an essential tool in drug
discovery, aids in predicting compound binding affinity with protein targets.
Therefore, this study aimed to analyze the interaction of Chalcomoracin, Morushalunin, and Guangsangon E with PD-1 and PPAR proteins, while also determining the physicochemical and
pharmacological properties of these compounds. To achieve this, an in silico test was conducted to assess interactions, using Autodock software and for visualization, the Discovery Studio was adopted.
2.1. The Biological Activity Prediction Using PASS
The 3 isolated compounds from
Morus sp, namely Chalcomoracin, Morushalunin, and Guangsangon E, obtained
through tissue culture, were analyzed for their biological activity using the
Prediction of Activity Spectra for Substances (PASS). The online platform,
accessible at http://www.pharmaexpert.ru/passonline, was used for this purpose.
Canonical SMILES of each compound were inputted on the website and a list of
the biological activity was generated based on the existing database on PASS.
The results included Pa (Probably active) and Pi (Probably active) values.
Finally, when the Pa and Pi values are closer to 1 and 0, respectively, it
signified better and good performance.
2.2. Prediction of Pharmacological Activity (ADME) of Compounds
and Drug Likeness Score
ADME characteristics of the
compounds were analyzed using SwissADME (http://www.swissadme.ch/). Canonical
SMILES of each compound were incorporated into Swiss ADME. Swiss ADME,
providing a predicted pharmacological profile. Additionally, https://www.molinspiration.com/
and https://molsoft.com/mprop/ were constituted to verify compliance with
Lipinski's rules.
2.3. Molecular Docking
The proteins used were Human PD-1
(PDB ID: 57w9) and PPAR(PDB ID: 5two), sourced from the RCSB Protein Data
Bank (https://www.rcsb.org/). Removal of unnecessary water molecules, ligands,
and chains was conducted. Autodock Vina 1.5.7 served as the docking software.
Validation of grid box dimensions was performed, with the dimensions for PD-1
being x= 50, y=50, and z = 50, centered at x= 14.974 Å, y= 30.713 Å, z= 187.813
Å, and for PPAR
x= 40, y=40, and z = 40, centered at x= -23.939 Å, y= -20.434
Å, z= 9.727 Å. The grid box dimensions were selected based on the RMSD value
(Sahlan et al., 2020). After docking, visualization was performed using
Discovery Studio.
Figure 2 Research Frameworks of This Study
3.1.
Prediction of the Biological Activity of
Compounds
A computer program called PASS,
accessible at (http://www.pharmaexpert.ru/passonline/) was used to predict bioactivity
spectra based on chemical structures. This computational method facilitated
potential in vivo
bioactivity for
chalcomoracin, guangsangon E, and morushalunin. Furthermore, this method produced a
comprehensive list of
biological activities along with their Pa and Pi. From the PASS analysis,
activities related to anticancer mechanisms were selected with a cut-off value
of >0.6. The selected activities include free radical scavenger, HIF-1a inhibitor,
apoptotic agonist, MMP9 Expression inhibitor, and chemopreventive. HIF-1a had a relationship with increased
PD-L1 during hypoxia. Additionally, it can increase PD-1 protein expression (Guo et al., 2022). Table 1 shows the result of the PASS analysis. Guangsangon E shows HIF-1a inhibitor effects, but its Pa value falls below Morushalunin. MMP-9, a
crucial element in
cancer metastasis, was influenced by chalcomoracin and guangsangon compounds,
indicating their activity on this protein.
Table 1 Biological
activity related to cancer prediction results analyzed using PASS.
|
Anticancer Activities |
Chalcomoracin |
Guangsangon E |
Morushalunin | |||
|
Pa |
Pi |
Pa |
Pi |
Pa |
Pi | |
|
Free Radical Scavenger |
0.620 |
0.005 |
0.631 |
0.005 |
- |
- |
|
HIF-1a inhibitor |
- |
- |
0.623 |
0.029 |
0.876 |
0.007 |
|
Apoptosis Agonist |
0.629 |
0.023 |
0.623 |
0.024 |
- |
- |
|
MMP-9 expression inhibitor |
0.623 |
0.013 |
0.678 |
0.008 |
- |
- |
|
Chemopreventive |
- |
- |
0.649 |
0.008 |
0.602 |
0.010 |
3.2.
Prediction of ADME of Compounds and Drug
Likeness Score
Bioavailability
Radar of SwissADME showed 6
physicochemical properties, including
lipophilicity, size, polarity, solubility, flexibility, and saturation.
The pink area represented the
optimal range for each property, which
comprises lipophilicity: XLOGP3 between -0.7 and +5.0, size: MW
between 150 and 500 g/mol, polarity: TPSA between 20 and 130 Å2, solubility:
log S not higher than 6, saturation: fraction of carbons in the sp3
hybridization not less than 0.25, and flexibility: no more than 9 rotatable
bonds (Daina, Michielin and Zoete,
2017). Based on the SwissADME bioavailability radar, it was observed that the 3 compounds have
poor bioavailability due to their
physico-chemical properties. According
to several studies, polyphenol indicated biological activity at low plasma concentrations. To enhance
the bioavailability of phenolic compounds, various methods were adopted, such as modifying the formulation or engaging in chemical derivatization. Curcumin is an example of a
beneficial polyphenol with poor bioavailability
Drug-likeness scores (DLS) from the 3 compounds were assessed using
the Molinspiration web server, as presented in Table 2. These scores
compared the physicochemical properties of the compounds with those of existing drugs based on
Lipinski’s rules. DLS
usually ranged from 0 to 1, where a score of 1 indicated a good candidate for drug development. Conversely, a score of 0 implies that the compound is less likely to be a drug (Sampat et al., 2022). In the context of this study, a DLS score
above 0 was observed. This information is valuable in predicting
whether the compound can be synthesized or evaluated.
Figure
3 Bioavailability
Radar of SwissADME analysis (a) Chalcomoracin, (b) Guangsangon E, (c)
Morushalunin
Table 2 ADME of Compounds and Drug
Likeness Score
|
No |
Compounds Name |
Software |
MW |
Log P |
TPSA* (A2) |
HBD |
HBA |
Rotatable Bond |
DLS |
|
1 |
Chalcomoracin |
SwissADME |
648.70 |
8.26 |
171.82 |
7 |
9 |
7 |
1.14 |
|
|
|
Molsoft.com |
648.24 |
8.86 |
138.28 |
- |
| ||
|
|
|
Molinspiration cheminformatic |
648.71 |
8.98 |
171.81 |
- |
| ||
|
2 |
Guangsangon E |
SwissADME |
648.70 |
8.26 |
171.82 |
7
|
9 |
7 |
1.09 |
|
Molsoft.com |
648.24 |
8.83 |
139.35 |
- |
| ||||
|
Molinspiration cheminformatic |
648.71 |
8.79 | 171.81
|
- |
| ||||
|
3 |
Morushalunin |
SwissADME |
660.71 |
8.43 | 149.82 |
5 |
9 |
6 |
0.79 |
|
Molsoft.com |
660.24 |
8.77 | 117.99 |
|
|
- | |||
|
Molinspiration cheminformatic |
660.72 |
8.97 | 149.82 |
|
|
- |
Notes: MW: Molecular Weight; TPSA: The Polar Surface Area; HBD: Hydrogen Bond Donor; HBA: Hydrogen Bond
Acceptor; DLS: Drug Likeness Score
3.3. Molecular
Docking
Molecular docking was conducted
to explore the potential interactions between chemical compounds derived from Morus sp and the PD-1 and PPAR proteins. Furthermore, it is a computational method aimed at identifying ligands that are
geometrically and energetically suitable for a given receptor (Suhartanto et al., 2017). Autodock Vina
software was chosen for this analysis, as previous
studies have showed its superior
accuracy compared to other options such as PatchDoc (Sahlan et al. 2023).The critical residues included in the PD-1/PD-L1 interaction were VAL64, ILE126, LEU128, ALA132,
ILE134, ILE54, TYR56, MET115, ALA121, and TYR123. The result shows
chalcomoracin was bound to protein through 4 hydrogen
bonds at amino acid residues such as ALA 121, ASP 122, SER 17, and TYR 123. Guangsangon showed hydrogen binding on ALA121, ILE54, and TYR123, while morushalunin interacted
with ARG 125, ASP 122, and TYR
123. The 3 compounds had protein interaction
through hydrogen bonds on several critical residues in the PD-1/PDL-1
interaction, as detailed in Table 3. They can bond with the TYR 123 amino acid residue. Morushalunin had the
lowest
compared to chalcomoracin and guangsangon E. Additionally, it had the highest inhibition constant (Ki) value.
Figure 4 Compounds Three Dimension Structures (a) Chalcomoracin, (b)
Guangsangon E, (c) Morushalunin
Tables 3 Molecular Docking Results of Chalcomoracin, Guangsangon E, and
Morushalunin on PD-1/PDL1 Protein
|
Compounds Name |
|
Inhibition Constanta (Ki) |
Hydrogen Bond |
|
Chalcomoracin |
-6.22 |
27.41 µm |
ALA 121, ASP 122, SER 17, TYR 123 |
|
Guangsangon E |
-8.91 |
292.2 nm |
ALA121, ILE54, TYR123 |
|
Morushalunin |
-9.28 |
157.41 nm |
ARG 125, ASP 122,TYR 123 |
Phenolic compounds such as chalcomoracin, guangsangon E, and chalcomoracin can form
complexes with protein through covalent or non-covalent interaction, hydrogen, van der Waals,
electrostatic, and
hydrophobic bonding. The primary modes of interaction are
predominantly hydrophobic
interaction and hydrogen binding (Shahidi and
Dissanayaka, 2023).
Hydrophobic
interaction between chalcomoracin and protein PD-1/PDL-1 occurs through
pi-alkyl binding with amino acid residue MET115, ILE54, ALA121, and TYR56.
Additionally, there was evidence of pi-cation interaction comprising LYS124 and
ASP 122, which showed a high strength compared to hydrogen
bonds. A P-alkyl binding pattern was identified between morushalunin and
PD-1/PDL-1, interacting with residues MET115, ILE54, ALA121, and TYR56 similars
to chalcomoracin. In the case of guangsangon, pi-alkyl interaction was
specifically observed with residue ALA18.
|
|
|
|
|
Figure 5 Two-Dimensional (2D) Visualizations Interaction with Protein PD-1 (a) Chalcomoracin, (b) Guangsangon
E, (c) Morushalunin
Table 4 Molecular Docking Results of
Chalcomoracin, and Guangsangon E on PPAR
|
Compounds Name |
|
Inhibition Constant |
Hydrogen Bond |
|
Chalcomoracin |
-5.69 |
67.39 µm |
ARG 280, CYS 285, SER299, TYR327, |
|
Guangsangon E |
-12.29 |
977.08 pm |
GLU343, GLY284, HIS449 |
This
study showed that Guangsangon E, with its lower and Ki
values, were the superior ligand for PPAR
. This is supported by its more
spontaneous interactions and the formation of more stable protein-ligand
complexes, as evidenced in Table 4.
Figure 6
Two-Dimensional
(2D) Visualizations Interaction with Protein PPAR (a) Chalcomoracin, (b)
Guangsangon E
In conclusion, the PASS analysis indicated that
Morushalunin and Chalcomoracin had activity as HIF1a and MMP-9 expression
inhibitors, respectively. Meanwhile, Guangsangon E showed activity on both
proteins. DLS for Chalcomoracin, Morushalunin, and Guangsangon E were 1.14,
1.09, and 0.79 respectively. According to the SwissADME bioavailability radar,
all three compounds demonstrated poor bioavailability due to their
physicochemical properties. It is important to note that several polyphenols
manifested biological activity at low plasma concentrations. To
address the issue of poor bioavailability, various strategies were adopted,
such as modifying the formulation or chemical derivatization. In this study, the interaction between Guangsangon E and PPAR-y was -12.29
kcal/mol, while of Chalcomoracin with PPAR-y was -5.69 kcal/mol. The docking
scores for Chalcomoracin, Morushalunin, and Guangsangon E on PD-1 proteins were
-6.21 kcal/mol, -8.91 kcal/mol, and -9.28 kcal/mol, respectively. Both Chalcomoracin and Guangsangon E
are bound to PD-1 and PPAR-
suggesting potential significance in breast
cancer pathogenesis. On the other hand, Morushalunin exclusively interacted
with the PD-1 protein. These indicated the potential of the compound to relate
with PD-1 and PPAR
key proteins in breast cancer pathogenesis. Therefore,
further study is needed to validate these predictions.
The authors are grateful to the
Directorate of Research and Development, Universitas Indonesia under HIBAH PUTI
2022 Grant No. NKB-1248/UN2.RST/HKP.05.00/2022, for funding this study.
| Filename | Description |
|---|---|
| R2-CE-6707-20230929234224.png | --- |
| R2-CE-6707-20230929234245.png | --- |
| R2-CE-6707-20230929234257.png | --- |
| R2-CE-6707-20230929234312.png | --- |
| R2-CE-6707-20230929234332.png | --- |
| R2-CE-6707-20230929234346.png | --- |
| R2-CE-6707-20230929234358.png | --- |
| R2-CE-6707-20230929234413.png | --- |
Abourashed,
E., 2013. Bioavailability of Plant-Derived Antioxidants. Antioxidants, Volume
2(4), pp. 309–325
Daina, A., Michielin, O., Zoete, V., 2017. SwissADME: a Free Web Tool to
Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of
Small Molecules. Scientific Reports,
Volume 7(1), p. 42717
Fitriani, R.,
Happyana, N., Hakim, E.H., 2021. Potential Cytotoxic Diels-Alder Type Adducts
From the Liquid Medium of Morus Alba Var. Shalun Root Cultures. Natural Product Research,
Volume 35(13), pp. 2274–2278
Giaquinto, A.N.,
Sung, H., Miller, K.D., Kramer, J.L., Newman, L.A., Minihan, A., Jemal, A.,
Siegel, R.L., 2022. Breast Cancer Statistics, 2022. A Cancer
Journal for Clinicians, Volume 72(6), pp. 524–541
Guo, T., Wang, T.,
Zhang, J., Chen, S., Wang, X., 2022. HIF1A Predicts the Efficacy of Anti-PD-1
Therapy in Advanced Clear Cell Renal Cell Carcinoma. Translational Oncology, Volume 26, p. 101554
Lukasiewicz, S.,
Czeczelewski, M., Forma, A., Baj, J., Sitarz, R., Stanislawek, A., 2021. Breast
Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and
Current Treatment Strategies—An Updated Review. Cancers,
Volume 13(17), p. 4287
Janakirama, A.R.R.S.,
Shivayogi, S.M., Satyanarayana, J.S., Kumaran, R.C., 2020. Characterization of
Isolated Compounds from Morus Spp. and Their Biological Activity as Anticancer
Molecules. BioImpacts, Volume 11(3), pp.
187–197
Sahlan, M., Dewi,
L.K., Pratami, D.K., Lischer, K., Hermansyah, H., 2023. In Silico
Identification of Propolis Compounds Potential as COVID-19 Drug Candidates
Against SARS-CoV-2 Spike Protein. International Journal of
Technology, Volume 14(2), p. 387
Sahlan, M., Faris,
M.N.H. Al, Aditama, R., Lischer, K., Khayrani, A.C., Pratami, D.K., 2020. Molecular
Docking of South Sulawesi Propolis against Fructose 1,6-Bisphosphatase as a
Type 2 Diabetes Mellitus Drug. International Journal of
Technology, Volume 11(5), p. 910
Sampat, G.,
Suryawanshi, S.S., Palled, M.S., Patil, A.S., Khanal, P., Salokhe, A.S., 2022. Drug
Likeness Screening and Evaluation of Physicochemical Properties of Selected
Medicinal Agents by Computer Aided Drug Design Tools. Advances in Pharmacology and Pharmacy,
Volume 10(4), pp. 234–246
Schütz, F., Stefanovic,
S., Mayer, L., Au, A. von, Domschke, C., Sohn, C., 2017. PD-1/PD-L1 Pathway in
Breast Cancer. Oncology Research and
Treatment, Volume 40(5), pp. 294–297
Shahidi, F.,
Dissanayaka, C.S., 2023. Phenolic-Protein Interactions: Insight from In-Silico
Analyses – A Review. Food Production,
Processing and Nutrition, Volume 5(1), p. 2
Suhartanto, H.,
Pasaribu, A.P., Siddiq, M.F., Fadhila, M.I., Hilman, M.H., Yanuar, A., 2017. A
Preliminary Study on Shifting from Virtual Machine to Docker Container for
Insilico Drug Discovery in the Cloud. International
Journal of Technology, Volume 8(4), p. 611
Sung, H., Ferlay,
J., Siegel, R.L., Laversanne, M., Soerjomataram, I., Jemal, A., Bray, F., 2021.
Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality
Worldwide for 36 Cancers in 185 Countries. A Cancer
Journal for Clinicians, Volume 71(3), pp. 209–249
Wang, Y., Zhu, M.,
Yuan, B., Zhang, K., Zhong, M., Yi, W., Xu, X., Duan, X., 2018. VSP-17, a New
PPAR Agonist, Suppresses the Metastasis of Triple-Negative Breast Cancer via
Upregulating the Expression of E-Cadherin. Molecules,
Volume 23(1), p. 121
Yang, Y., Tan, Y.-X., Chen, R.-Y., Kang, J., 2014. The Latest Review on The Polyphenols And Their Bioactivities of Chinese Morus Plants. Journal of Asian Natural Products Research, Volume 16(6), pp. 690–702Yang, Y., Tan, Y.-X., Chen, R.-Y., Kang, J., 2014. The Latest Review on The Polyphenols And Their Bioactivities of Chinese Morus Plants. Journal of Asian Natural Products Research, Volume 16(6), pp. 690–702
