Association between ADIPOQ (rs1501299) SNP with Insulin Resistance in Indonesian Type 2 Diabetes Mellitus Patients
Corresponding email: donny.nauphar@ui.ac.id
Published at : 07 Dec 2023
Volume : IJtech
Vol 14, No 7 (2023)
DOI : https://doi.org/10.14716/ijtech.v14i7.6691
Nauphar, D., Alfaqih, R.I.M., Brajadenta, G.S., Pratamawati, T.M., 2023. Association between ADIPOQ (rs1501299) SNP with Insulin Resistance in Indonesian Type 2 Diabetes Mellitus Patients. International Journal of Technology. Volume 14(7), pp. 1578-1585
| Donny Nauphar | Department of Genetics, Faculty of Medicine, Universitas Swadaya Gunung Jati, Cirebon, West Java, 45132, Indonesia |
| Robby Irham Maulana Alfaqih | Bachelor of Medicine, Faculty of Medicine, Universitas Swadaya Gunung Jati, Cirebon, West Java, 45132, Indonesia |
| Gara Samara Brajadenta | Department of Genetics, Faculty of Medicine, Universitas Swadaya Gunung Jati, Cirebon, West Java, 45132, Indonesia |
| Tiar M Pratamawati | Department of Genetics, Faculty of Medicine, Universitas Swadaya Gunung Jati, Cirebon, West Java, 45132, Indonesia |
Insulin resistance is an
important aspect of metabolic endocrine disorder, and adiponectin functions as
an insulin-sensitizer. Changes in adiponectin levels are associated with
alterations in insulin sensitivity. Insulin resistance results from various
variables that contributes to abnormalities in insulin signaling, including a
decrease in adiponectin levels. Genetics is recognized as one of the key
elements influencing adiponectin levels, with investigations showing that
ADIPOQ SNP can impact insulin sensitivity and plasma adiponectin levels.
Therefore, this study aimed to examine association between ADIPOQ gene
polymorphism in patients with type 2 diabetes mellitus (DM) and insulin
resistance level. A case-control study was conducted with 60 participants
recruited from Sunyaragi Community Health Center in Cirebon, West Java. Data
were collected using fasting blood glucose (mg/dl) and Polymerase Chain
Reaction – Restriction Fragment Length Polymorphism (PCR-RFLP). The results
showed that the genotype frequency of SNP in the case group was GG = 12 (40%),
GT = 16 (53.33%), TT = 2 (6.67%). Meanwhile, in the control group, it was
observed to be GG = 18 (60%), GT = 11 (36.67%), and TT = 1 (3.33%).
Statistically analysis showed a significant association between +276 G/T polymorphism
and type 2 DM. This concluded that individuals with polymorphism are at higher
risk of developing type 2 DM.
Adiponectin; ADIPOQ; Insulin Resistance; SNP
Insulin resistance is a pathological disorder affecting
insulin-dependent cells such as skeletal cells and adipocytes, leading to a
diminished response to normal levels of circulating insulin. This condition can
give rise to various health complications, including hyperglycemia,
hypertension, dyslipidemia, endothelial dysfunction, and metabolic disorders
such as metabolic syndrome or type 2 diabetes mellitus (DM)
Obesity is a risk factor for insulin resistance,
particularly in instances of excess fat accumulation. The metabolic effects
associated with insulin resistance serve as valuable clinical indicators for
identifying this condition
The main effect of insulin resistance is
development of type 2 DM. In this context, increased insulin production acts as
a compensatory mechanism, leading to insulin resistance. However, pancreatic
beta cells become damaged over time and are unable to meet insulin needs,
resulting in hyperglycemia. Insulin resistance also contribute to other
disorders such as metabolic syndrome, obesity, cardiovascular disease,
nonalcoholic fatty liver disease, and polycystic ovarian syndrome (PCOS)
According to the International Diabetes
Federation (IDF), an estimated 463 million individuals worldwide, aged 20 to
79, were diagnosed with DM in 2019. Southeast Asia
ranks third with a prevalence of 11.3%, and
Indonesia has approximately 1 million diabetics, as per the 2018 Basic Health
Research conducted by
The protein adiponectin, an insulin
sensitizer encoded by ADIPOQ gene and released by adipose cells, plays a
crucial role in reducing the rate of gluconeogenesis in the liver, enhancing
the absorption of glucose, and maintaining insulin sensitivity. Single
nucleotide polymorphisms, frequently called SNP, the most common type of
genetic variation, can alter the transcription rate of mRNA, influencing
protein production. SNP also has physiological impact on protein activity by
changing the nucleic acid, thereby altering the type of amino acid produced.
Insulin resistance can result from low plasma levels of the hormone
adiponectin, influenced by various factors such as genetics, diet, exercise,
and abdominal obesity
Study in diverse population showed that SNP of ADIPOQ gene could
influence the transcription rate or alter the amino acid sequence, consequently
affecting plasma adiponectin levels and insulin sensitivity. However, this
investigation have not been conducted in Indonesian. Despite that adiponectin operates as an insulin-sensitizer
indirectly, when it experience a drop in the levels, there will also be a
decrease in insulin sensitivity. This significantly plays a role in the
formation of insulin resistance among type 2 DM patients. By investigating the alleles, genotype, and
potential predisposition to type 2 DM in Indonesians, this study aims to
contribute insights that could inform preventive strategies.
In accordance with the previous explanation, the focus is specifically
on understanding how genetic factors, particularly ADIPOQ gene, affect insulin
resistance. The purpose of this study is to examine association between ADIPOQ
gene polymorphism in type 2 DM patients and insulin resistance levels in the Indonesian
population.
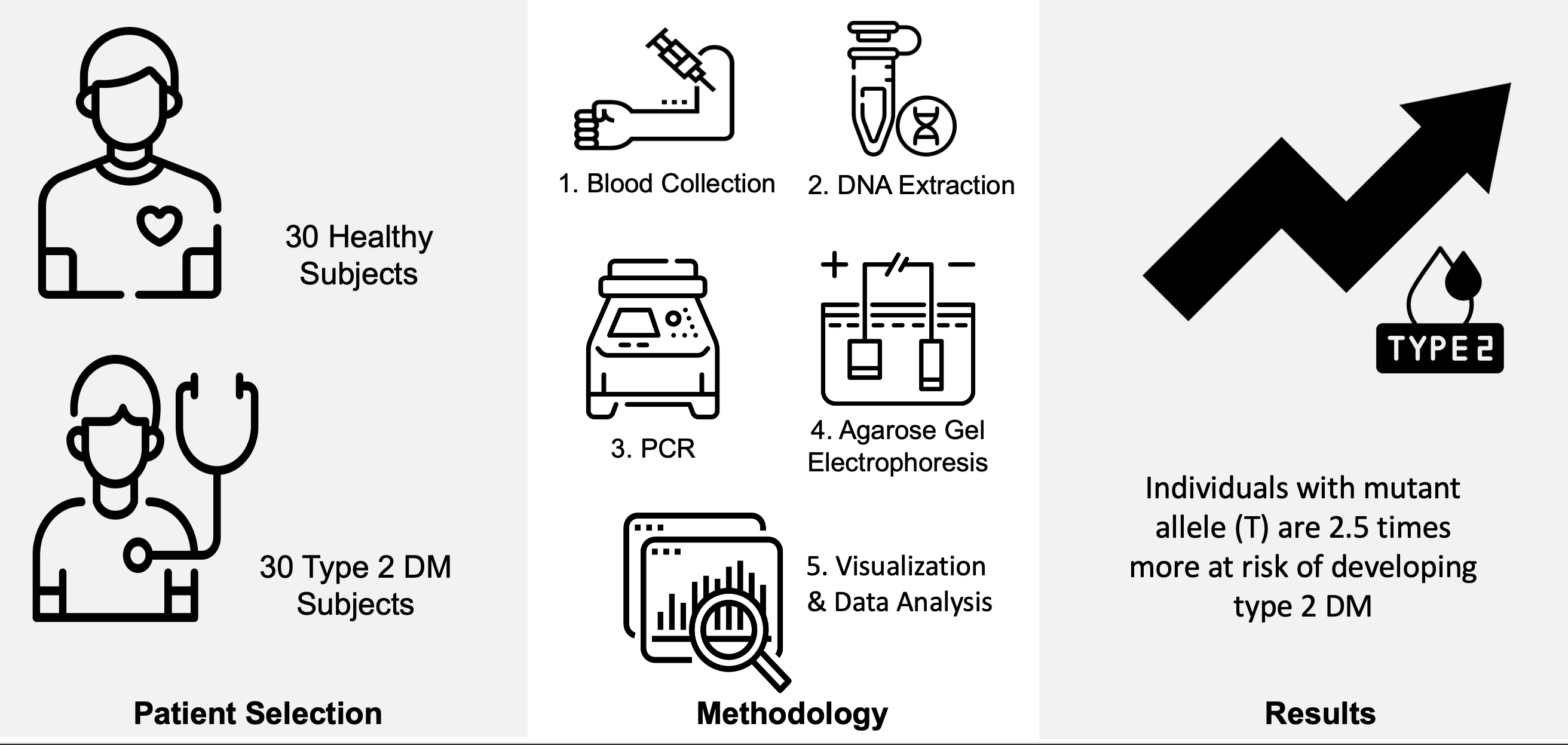
2.1. Patient Selection
This study was an analytical observational case-control investigation comprising 30 case and 30 control subjects. Ethical approval was received from the Medical Faculty Research Ethics Committee at Swadaya Gunung Jati University (131/EC/FKUGJ/V/2022). The investigation was conducted at the Sunyaragi Community Health Center in Cirebon, West Java and the Faculty of Medicine's Laboratory of Genetics and Molecular Biology. The control group consisted of individuals without type 2 DM diagnosis, while the cases group met PERKENI criteria for type 2 DM within a 3-month period. Exclusion criteria included type 1 DM, cancer, autoimmune illnesses, and subjects on steroid anti-inflammatory medication. After patients have fasted for 8 hours, the fasting blood glucose levels was determined through proper examination.
2.2. Nucleic Acid Extraction
After initial screening of medical
records and obtaining informed consent for sampling, 3 mL of peripheral blood
was drawn in EDTA for genetic analysis. The TianGen TIANamp Hi-DNA/RNA
Extraction Kit was used for blood extraction. The concentration and purity of
DNA were assessed using the Maestrogen MaestroNano Pro Spectrophotometer.
Finally, extracted DNA was stored at -20°C.
2.3. Genetic Analysis
DNA amplification was performed using BioRad T100 thermal cycler with forward primer: 5’-CCT GGT GAG AAG GGT GAG AA -3’ and reverse primer: 5’-AGA TGC AGC AAA GCC AAA GT- 3’. The amplification protocol included denaturation at 95? for 5 minutes, 35 cycles consisting of 95? for 30 seconds, 65? for 30 seconds, 72? for 30 seconds, 72? for 8 minutes, and ended with 25? for infinity hold. A 2% electrophoretic gel confirmed the 241 bp PCR product using the BioRad GelDoc EZ Imager, to ensure that DNA has been amplified. After amplification, the product was cut with the BsmI restriction enzyme. Restriction results were analyzed by a 2% agarose gel to observe RFLP. The expected outcomes on the agarose gel were GG (Homozygous Wildtype) genotype cut to 95bp and 146bp, GT (Heterozygote) cut to 95bp, 146bp, and 241bp, and TT (Homozygous Mutant) cut to 241bp.
2.4. Data Analysis
The frequency of each genotype was calculated and presented as percentage. To determine association between the independent and the dependent variable, an unpaired t-test of >2 groups was used to compare the genotype at SNP +276 with the average GDP level. Polymorphism at +276 G/T with type 2 DM was evaluated using a contingency test with a 2x2 table to derive odds ratio and p-value.
Figure 1 Schematic illustration of the workflow of the
study
3.1. Patient Characteristics
This present study
comprised 60 subjects, evenly divided into 30 cases and 30 controls, all
meeting the predefined inclusion and exclusion criteria. Both body height and weight were measured with
participants dressed in light clothing and without shoes. Obesity was assessed by body mass
index (BMI), calculated as weight in kilograms divided by the square of height in meters.
In the case group, 23
subjects fell under the normal body weight category, while 7 subjects were
classified as overweight. Approximately 66% of the samples in this group showed
increased fasting blood glucose levels. Meanwhile, the control had 10 overweights,
and all subjects showed elevated fasting glucose levels (Table 1).
Table 1 Subject Characteristics based on age, sex, weight, height, BMI, and
fasting glucose
|
|
Case (n) |
Control (n) |
% |
|
Age |
|
|
|
|
30-40 41-50 51-60 61-70 >71 |
1 5 9 10 5 |
5 10 5 5 5 |
10% 25% 23.3% 25% 16.7% |
|
Sex |
|
|
|
|
Male Female |
11 19 |
9 21 |
33.3% 66.7% |
|
Weight (kg) 45-50 51-60 61-70 71-80 81-85 |
3 8 10 8 1 |
1 8 17 4 0 |
6.7% 26.7% 45% 20% 1.6% |
|
Height (cm) 150-160 161-170 171-175 |
6 20 4 |
4 24 2 |
16.7% 73.3% 10% |
|
BMI Underweight Normal Overweight |
0 23 7 |
2 18 10 |
3.3% 68.4% 28.3% |
|
Fasting Glucose Normal Abnormal |
10 20 |
0 30 |
16.7% 83.3% |
3.2.
Genotypic Data
The results of PCR amplification are presented in
Figure 2 (a), showing a band at 241bp from L1-L3, which corresponded to control
sample 1-3 and non-template control (NTC) respectively. The results of the
restriction enzymes were shown in Figure 1b. Furthermore, the uncut band at
241bp showed the presence of G allele, while cut bands at 146bp and 95bp showed
the presence of T allele. In Figure 2 (b), subject K1 had bands at 241bp,
146bp, and 95bp, signifying the GT genotype.
Insulin resistance is an important
aspect of metabolic endocrine disorder. Adiponectin functions as an
insulin-sensitizer and changes in its level will also lead to alteration in
insulin sensitivity. Insulin resistance occured as a result of various factors
leading to abnormalities in insulin signaling, one of which is a decrease in
adiponectin levels. Genetics is one of the elements influencing adiponectin
levels. Several studies showed that ADIPOQ rs1501299 +276 G/T SNP increased the
possibility of developing type 2 DM and insulin resistance
Figure 2 (a) PCR amplification results and (b) Restriction Enzyme
digestion results. Figure 2(a) showed the PCR amplification result of 241bp
prior to enzyme restriction (L1, L2, L3) and a non-template control (NTC) was
added as a negative control. Figure 2(b) showed bands after restriction enzyme
digestion where K1-K14 are control cases. K1
showed bands of 241bp, 146bp, and 95bp, which means one allele was cut by the
restriction enzyme and the other was not, showing G and T allele and GT
genotype
Despite +276 G/T SNP being
a silent mutation, it converts G nucleotide base to T without altering the
amino acid composition of the protein generated by ADIPOQ gene. Studies showed
that ADIPOQ +276 G/T SNP affected adiponectin levels. Individuals with T allele
have lower adiponectin levels and index of insulin resistance, despite lack of
alteration in the protein
The results of this study
showed that the control had less polymorphism +276 G/T (21.7%), where the case
group had more at 33.3%, as presented in Table 2. Examining the genotype
frequencies of SNP +276 G/T in the case group indicated 12 (40%) GG, 16
(53.33%) GT, and 2 (6.67%) TT genotypes. Meanwhile, the control group had 18
(60%) GG, 11 (36.67%) GT, and 1 (3.33%) TT genotype, as presented in Table 3.
It was also discovered that the case group had a lower frequency of GG genotype
than the control, accompanied by a higher frequency of GT and TT genotypes.
Table 2 Allele frequency distribution
|
Allele |
Cases |
Control | ||
|
n |
% |
n |
% | |
|
G |
40 |
66.7% |
47 |
78.3% |
|
T |
20 |
33.3% |
13 |
21.7% |
|
Total |
60 |
100% |
60 |
100% |
Table 3 Genotype frequency distribution
|
Genotype |
Cases |
Control | ||
|
n |
% |
n |
% | |
|
GG |
12 |
40% |
18 |
60% |
|
GT |
16 |
53.33% |
11 |
36.67% |
|
TT |
2 |
6.67% |
1 |
3.33% |
|
Total |
30 |
100% |
30 |
100% |
The results are similar with those of
studies conducted in Chinese, Iranian, and Japanese population (Alimi, Goodarzi, and Nekoei, 2021). The analysis of SNP+276 G/T, with a p-value of
0.001, showed a significant association between SNP and the incidence of type 2
DM in Indonesian population. It was also shown that individuals with
polymorphism had a 2.5 times higher risk of developing type 2 DM, as shown in
Table 4.
3.3.
Genotypic and Phenotypic Correlation
Table 4 Genotypic
Association between ADIPOQ SNP and Type 2 DM.
|
Allele |
Case (n) |
Control (n) |
OR |
p-value |
|
GT/TT |
18 |
12 |
2.5 |
0.001 |
|
GG |
12 |
18 |
Table 5 Association between ADIPOQ SNP
and mean Fasting Blood Glucose Levels
|
Genotype |
Case (n) |
FBG ± SD (range)
(mg/dl) |
p-value |
|
GG |
12 |
124 ± 112
(110-342) |
0.054 |
|
GT TT |
16 2 |
230 ± 108
(116-460) 127 ± 26
(110-144) |
The results were not in line with studies in the Arab and Korean
population, where no association between SNP was identified, with a p-value of
0.69
No
significant differences were observed between the genotypes of +276 G/T
polymorphism in ADIPOQ gene in the case group with fasting blood glucose
levels. A p-value of 0.054 was discovered in the statistical analysis, showing
that there was no statistically significant difference between the 3 types of
genotypes examined (Table 5). In the study by
This
study has a primary limitation, stemming from the absence of plasma adiponectin
level data, which could influence insulin resistance levels. Furthermore, the
HOMA-IR examination, typically used for assessing type 2 DM levels, was not
applied in this investigation. Consequently, a direct link between SNP +276 G/T
and insulin resistance cannot be established. It is important to note that
other genes, such as GLUT4 and IRS, had stronger connections to insulin
resistance and type 2 DM.
In conclusion,
approximately 60% of participants in this study who had type 2 DM experienced a
polymorphism of +276 G/T. In the case group, the distribution of SNP +276 G/T
genotype was 12 (40%), 16 (53.33%), and 2 (6.67%) with GG, GT, and TT genotype,
respectively. In the control group, the breakdown was 18 (60%), 11 (36.67%),
and 1 (3.33%) for GG, GT, and TT genotype concerning SNP +276 G/T genotype.
Statistical analyses showed a significant association between the +276 G/T
polymorphism and type 2 DM. However, the odds ratio value suggested that
individuals with +276 G/T polymorphism were 2.5 times more easy to have
developed type 2 DM. To present a more comprehensive understanding of the
predisposed risk in the Indonesian population, future studies should include a
larger number of subjects in this demographic to determine allele frequencies
accurately.
The authors are
grateful to the Universitas Swadaya Gunung Jati 2022 Internal Research Fund for
funding this study.
Al-Daghri, N.M.,
Al-Attas, O.S., Alokail, M.S., Alkharfy, K.M., Hussain, T., Yakout, S.,
Vinodson, B., Sabico, S., 2012. Adiponectin Gene Polymorphisms (T45G and
G276T), Adiponectin Levels and Risk For Metabolic Diseases in an Arab
Population. Gene,
Volume 493(1), pp. 142–147
Alimi, M.,
Goodarzi, M.T., Nekoei, M., 2021. Association of ADIPOQ rs266729 and rs1501299
Gene Polymorphisms And Circulating Adiponectin Level with the Risk of Type 2
Diabetes in a Population of Iran: a Case-Control Study. Journal of Diabetes and Metabolic Disorders, Journal of
Diabetes & Metabolic Disorders, Volume 20(1),
pp. 87–93
Condorelli, R.A.,
Calogero, A.E., Di Mauro, M., La Vignera, S., 2017. PCOS and Diabetes Mellitus:
From Insulin Resistance to Altered Beta Pancreatic Function, a Link in
Evolution. Gynecological Endocrinology,
Volume 33(9), pp. 665–667
De Luis, D.A.,
Izaola, O., De La Fuente, B., Primo, D., Ovalle, H.F., Romero, E., 2017.
Rs1501299 Polymorphism in the Adiponectin Gene and Their Association with Total
Adiponectin Levels, Insulin Resistance and Metabolic Syndrome in Obese Subjects.
Annals of Nutrition and Metabolism,
Volume 69(3–4), pp. 226–231
Frankenberg, A.D.V.,
Reis, A.F., Gerchman, F., 2017. Relationships Between Adiponectin Levels, the
Metabolic Syndrome, and Type 2 Diabetes: A Literature Review. Archives of Endocrinology and Metabolism,
Volume 61(6), pp. 614–622
Ministry of Health
Republic Indonesia,
2018. Basic Health Research 2018. Ministry of Health Republic Indonesia
Moon, H.U., Ha,
K.H., Han, S.J., Kim, H.J., Kim, D.J., 2014. Adiponectin, Visceral Fat in
Insulin Resistance and Secretion. Endocrinology, Nutrition
Metabolism, Volume 34(1), pp. 1–12
Nam, J.S., Han,
J.W., Lee, S.B., You, J.H., Kim, M.J., Kang, S., Park, J.S., Ahn, C.W., 2018. Calpain-10
and Adiponectin Gene Polymorphisms in Korean Type 2 Diabetes Patients. Endocrinology and Metabolism,
Volume 33(3), pp. 364–371
Nauphar, D.,
Wahidiyat, P.A., Ariani, Y., 2022. Molecular
Study in Identifying Genotypes to Phenotypes Relations of Transfusion-Dependent
Thalassemia Patients in Cirebon, West Java. International
Journal of Technology, Volume 13(8), pp. 1726–1734
Prakash, J.,
Mittal, B., Awasthi, S., Srivastava, N., 2015. Association of Adiponectin Gene
Polymorphism with Adiponectin Levels and Risk for Insulin Resistance Syndrome. International Journal of Preventive Medicine,
Volume 8(6), p. 31
Pratamawati, T.M.,
Alwi, I., Asmarinah., 2022. Methylenetetrahydrofolate Reductase (MTHFR) C677T
and A1298C Gene Polymorphism as Risk Factors for Essential Hypertension. International Journal of Technology,
Volume 13(8), pp. 1622–1629
Samuel, V.T.,
Shulman, G.I., 2016. The Pathogenesis of Insulin Resistance: Integrating
Signaling Pathways And Substrate Flux. Journal of
Clinical Investigation, Volume 126(1), pp. 12–22
Sung, K.C., Lee,
M.Y., Kim, Y.H., Huh, J.H., Kim, J.Y., Wild, S.H., Byrne, C.D., 2018. Obesity
and Incidence of Diabetes: Effect of Absence of Metabolic Syndrome, Insulin
Resistance, Inflammation And Fatty Liver. Atherosclerosis,
Volume 275, pp. 50–57
Yaribeygi, H.,
Farrokhi, F.R., Butler, A.E., Sahebkar, A., 2019. Insulin Resistance: Review of
the Underlying Molecular Mechanisms. Journal of Cellular
Physiology, Volume 234(6), pp. 8152–8161
Yu, K.T., Maung,
K.K., Thida, A., Myint, T., 2018. Single Nucleotide Polymorphism at +276 g>T
of the Adiponectin Gene and Plasma Adiponectin Level in Myanmar Type 2 Diabetic
Patients. Journal of the ASEAN Federation of Endocrine
Societies, Volume 33(2), pp. 160–164
Zhao, F., Mamatyusupu, D., Wang, Y., Fang, H., Wang, H., Gao, Q., Dong, H., Ge, S., Yu, X., Zhang, J., Wu, J., Song, W., Wang, W., 2016. The Uyghur Population and Genetic Susceptibility To Type 2 Diabetes: Potential Role for Variants in CAPN10, APM1 and FUT6 Genes, Journal of Cellular and Molecular Medicine, Volume 20(11), pp. 2138–2147
Ziemke, F., Mantzoros, C.S., 2010. Adiponectin in Insulin Resistance: Lessons From Translational Research. American Journal of Clinical Nutrition, Volume 91(1), pp. 258–261
