Molecular Study in Identifying Genotypes to Phenotypes Relations of Transfusion-Dependent Thalassemia Patients in Cirebon, West Java
Published at : 30 Dec 2022
Volume : IJtech
Vol 13, No 8 (2022)
DOI : https://doi.org/10.14716/ijtech.v13i8.6136
Nauphar, D., Wahidiyat, P.A., Ariani, Y., 2022. Molecular Study in Identifying Genotypes to Phenotypes Relations of Transfusion-Dependent Thalassemia Patients in Cirebon, West Java. International Journal of Technology. Volume 13(8), pp. 1726-1734
| Donny Nauphar | 1. Doctoral Program in Biomedical Sciences, Faculty of Medicine, Universitas Indonesia, Kampus UI Salemba, 10430, Indonesia, 2. Department of Genetics, Faculty of Medicine Universitas Swadaya Gunung J |
| Pustika Amalia Wahidiyat | Department of Pediatrics, Faculty of Medicine, Universitas Indonesia, Kampus UI Salemba, 10430, Indonesia |
| Yulia Ariani | Department of Pediatrics, Faculty of Medicine, Universitas Indonesia, Kampus UI Salemba, 10430, Indonesia |
Molecular characterization is
important prior to clinical management as it can provide additional information
for the clinical management of patients. This study aims to characterize the
most common mutation and identify genotype-to-phenotype relations in transfusion-dependent
thalassemia patients. A total of 30 transfusion-dependent patients were
recruited who had never undergone thalassemia detection. Peripheral blood
samples were collected and analyzed for hematological parameters, hemoglobin
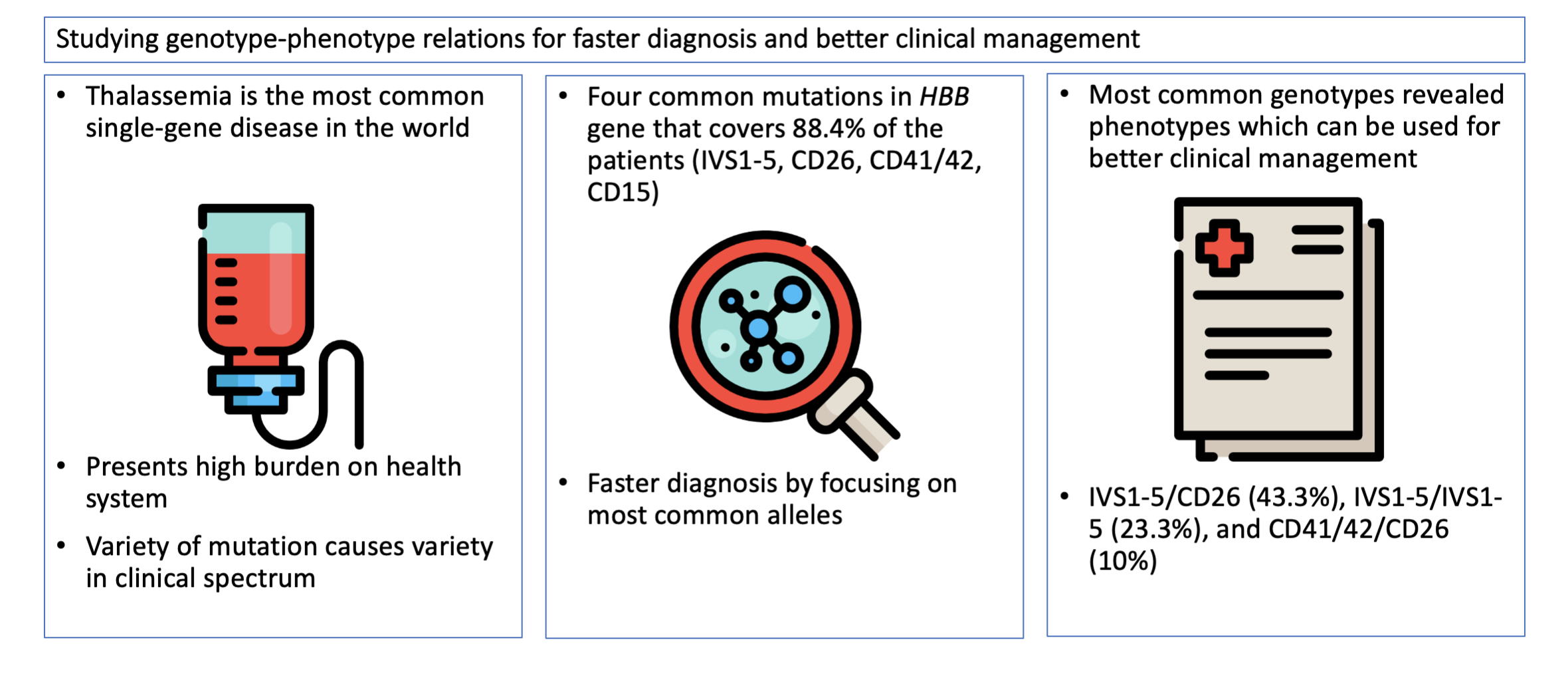
component, and HBA and HBB gene mutation analysis. The most
common mutation in the HBB gene was IVS1-5 (41.7%) and CD26/HbE (23.3%),
with homozygous IVS1-5 (23.3%) and IVS1-5/HbE (30%) as the most common
genotype. The study revealed a genotype and phenotype correlation of the most
common thalassemia mutations in Cirebon, West Java, Indonesia, with four
alleles dominating the genotype, covering 88.4% of the population. A
significant difference in HbA2+HbE and HbF levels was observed between
homozygous ?-thalassemia and
?-thalassemia/HbE. Homozygous ?-thalassemia with ?-thalassemia trait has better
hematological parameters compared to homozygous ?-thalassemia and ?-thalassemia/HbE but does not translate to a better
severity index. Characterizing the most common thalassemia mutations in
the Indonesian population can streamline the subsequent diagnostic approaches
by focusing on the small range of predominant alleles instead of a wide range
of alleles, which can provide critical data for better patient management.
Genotype-phenotype; HBA; HBB; Mahidol severity index; Thalassemia
Thalassemias are autosomal
recessive disorders marked by quantitative defects in globin chain synthesis.
This disease can be further classified based on the globin chain(s) affected by
defects in synthesis, whereby defects in the -globin chain cause
-thalassemia
while defects in the
-globin chain cause
-thalassemia (Nienhuis & Nathan, 2012; Piel & Weatherall,
2014).
Thalassemia is one of the most common inherited
hemoglobin disorders globally (Taher et al., 2018) and is currently a public health concern affecting Indonesia's
multi-
In thalassemia, emphasis should
be given to the severity of the clinical appearance and the complexity of the
genetic background associated with the disease. More than 100 types of -thalassemia with over 800 types of mutations and structural variants of the
-globin gene have been identified. And 300 types of ?-thalassemia with over
900 types of mutations and structural variants of the ?-globin gene have been
reported that lead to various clinical states because of the varying arrangements of compound
heterozygous alleles (Giardine et al., 2014; Farashi & Harteveld, 2018;
Thein, 2018). Clinical features of these conditions reflect a wide range of
transfusion requirements ranging from transfusion-independent to lifelong,
regular blood transfusions (Viprakasit & Ekwattanakit, 2018).
Thalassemia screening in
Indonesia relies on general practitioners to promptly identify the possibility
of thalassemia or hemoglobinopathy. And the traits while ruling out iron
deficiency as the first differential diagnosis. Identifying microcytic anemia
using blood analyzers and utilizing the Mentzer index were used as early
screening. Once referred to specialist centers, the diagnosis is usually
followed up with hemoglobin analysis and DNA analysis, although both services
are only available in larger cities and remain scarce in smaller cities and
rural areas (Wahidiyat et
al., 2020). Diagnosed patients receive blood transfusion according to their
hemoglobin levels (usually <7.0 g/dL) on an average of every 2-4 weeks,
depending on the need to maintain hemoglobin levels at 9-10.5 g/dL. Patients
also receive iron chelation therapy to treat iron overload and prevent cardiac
hemosiderosis.
Since the diversity of the
clinical spectrum is closely related to the variety of mutations, it is
important to perform molecular characterization of the genotype to gain
information on disease severity, expected transfusion requirement, and
chelation therapy before clinical management for thalassemia. In current
existing clinical setups in Indonesia, most diagnoses of thalassemia are based
on data obtained from clinical findings, erythrocyte morphology, and hemoglobin
levels rather than molecular characterization.
Therefore, this study aims to
investigate the genotype-to-phenotype relationship of these mutations by
studying the hematological parameters, hemoglobin typing, and disease severity.
This study will show the most common mutations, allelic and genotypic
frequency, and genotype-phenotype correlation of the mutations as disease
severity of transfusion-dependent patients enrolled at Cirebon, West Java,
Indonesia.
2.1. Patient Selection
Thirty transfusion-dependent thalassemia patients were
enrolled at the Hematology, Oncology, and Thalassemia Clinic, Gunung Jati
Regional Public Hospital in Cirebon. None of the patients have previously
undergone molecular analysis for thalassemia detection. Written informed
consent was obtained from all participants prior to enrolment. A written
consent was obtained from their parents or guardians for underaged patients.
The Institutional Review Board of Faculty of Medicine Universitas Swadaya
Gunung Jati, Cirebon, Indonesia, approved the study protocol and followed the
ethical principles of the Declaration of Helsinki of 1975 and its revision. All
patients were previously classified as thalassemic based on hemoglobin levels
and clinical appearance. There were 12 males and 18 females, with an average
age of 12.77 years. Patient clinical history was recorded along with the age of
onset, age at first blood transfusion, transfusion requirements, spleen size,
height, and weight.
2.2. Hematological and Biochemical
Analysis
Hematological analysis was
carried out using Sysmex XN-1000 Automated Hematology Analyzer (Sysmex
Corporation, Kobe, Japan). Hemoglobin component analysis was determined using
the BioRad Variant II HPLC system (Bio-Rad Laboratories Inc, Hercules, CA, USA)
according to the manufacturer’s instructions.
2.3. Genetic Analysis
Peripheral
blood samples were collected in EDTA. Genomic DNA extraction was carried out
using the spin-column method. Mutations in the HBA
gene were analyzed using multiplex GAP-PCR which detects –3.7
(NG_000006.1: g.34164_37967del3804), –
4.2, --SEA
(NG_000006.1:g.26264_4556 4del19301), --THAI (NG_
000006.1:g.10664_44164del33501) mutations as previously described (Chong et al., 2000a; Chong et
al., 2000b). The mutations in
the HBB gene were analyzed using
the Reverse Dot Blot method to simultaneously detect the 10 most common Thai HBB gene mutations [-28
(A>G), CD17 (HBB:c.52A>T); CD19 (HBB:c.59A>G), CD26 (HbE)
(HBB:c.79G>A), IVS1-1
(G>T) (HBB:c.92 + 1G>T), IVS1-5 (G>C) (HBB:c.92 + 5G>C); CD35
(HBB:c.35C>A), CD41/42 (HBB:c.127_130delCTTT), CD71-72 (HBB:c.216_217insA),
and IVS2-654 (HBB:c.316-197C>T)]
(Sutcharitchan et al., 1995).
2.4. Disease Severity
The disease severity was classified into mild, moderate, or severe using the Mahidol Severity Index by measuring the hemoglobin level at steady state, age at thalassemia presentation, age at receiving a first blood transfusion, requirement for blood transfusion, size of the spleen, and growth and development (Sripichai et al., 2008).
Figure 1 Workflow schematic
3.1. Patient
Characteristics
The thalassemia patients in this study have an average age of 12.77 years old 20 of them have an age of onset below three years old (67%), and 23 subjects have an age of first blood transfusion below three years old (77%) as shown in Table 1.
Table 1 Characteristics of
transfusion-dependent thalassemia patients in Cirebon
|
|
|
Frequency |
% |
|
Average age (years) |
12.77 (± 6.99) |
|
|
|
Sex |
Male Female |
12 18 |
40% 60%
|
|
Age at onset |
<3 years old 3-10 years old >10 years old |
20 9 1 |
67% 30% 3%
|
|
Age at first blood transfusion |
<5 years old 5-10 years old >10 years old |
23 6 1 |
77% 20% 3%
|
|
Transfusion requirement |
>2x/year 1-2x/year <1x/year |
30 0 0 |
100% 0% 0%
|
|
Spleen size |
<3 cm 3-10cm >10cm Splenectomized |
15 14 1 0 |
50% 47% 3% 0%
|
|
Severity Index |
Normal Moderate Severe |
0 8 22 |
0% 26.7% 73.3% |
All
the patients underwent a hyper-transfusion regime, receiving more than two
blood transfusions in a month. The transfusion regime aims to keep hemoglobin
levels above ten g/dL. The hyper-transfusion regime, however, has a long-term
side effect of iron overload. Iron is required for viability and utilized in
heme formation and is central in functioning hemoglobins, the essential
oxygen-carrying molecule in all vertebrates. In contrast, free iron is
extremely cytotoxic as it can form reactive superoxide radical (O2-)
or hydrogen peroxide, generating the hydroxyl (•OH) radical. Which can oxidize
biological macromolecules. Including lipids, proteins, and DNA leading to
cellular damage (Leecharoenkiat et
al., 2016). Humans carry around 3-5 g
of iron in various forms, and since iron has low bioavailability, humans have
evolved to be highly efficient in conserving iron with no mechanism of
excreting excess iron under conditions of iron overload (Lawen & Lane,
2012). Humans only excrete 1-2 mg of iron daily,
which is less than 0.1% of the body's total iron, which is replenished through
dietary sources. Meanwhile, a unit of transfused red blood cells contains
around 200mg of iron, and in the absence of iron chelation, the iron will
continuously accumulate and result in iron overload (Nienhuis & Nathan,
2012). Iron overload, in turn, will lead to
irreversible organ damage, such as cirrhosis, diabetes, heart disease, and hypogonadism
(Leecharoenkiat et
al., 2016).
To
counter the effect of iron overload, thalassemia patients rely heavily on the
continuous use of iron chelators to aid the excretion of excess iron. There are
three significant iron chelators mainly used in Indonesia, namely deferoxamine
(DFO), deferiprone (DFP), and deferasirox (DFX) (Wahidiyat et al., 2020). Though these drugs are
widely available, the distribution and continuous supply of these drugs remain
a problem in smaller cities.
In
terms of disease severity, most patients fall under the severe phenotype base
in the Mahidol Severity Index (73.3%), followed by moderate severity (26.7%),
with no patients having the normal phenotype. This clinically means that all
subjects have to be monitored for their iron chelation therapy, growth
retardation, and spleen and liver enlargement to ensure a higher quality of
life.
3.2. Genotypic
Data
The
HBA mutation analysis (Table
2) showed 28 normal HBA with two patients with -3.7 mutation,
making -
3.7 the only mutation detected in the patient population.
The HBB
mutation analysis (Table 3) yielded eight alleles and nine genotypes. The most
common allele was IVS1-5 (46.7%), a mutation in the consensus splice site in
the first intron causing reduced splicing at the mutated donor site (Malik &
Tisdale, 2017), followed by hemoglobin E,
a mutation in codon 26 of HBB (31.7%), CD41/42 (5%), and CD15 (5%) respectively.
This finding is similar to previous studies conducted in Indonesia for HBB mutations, where IVS1-5
and HbE were generally found throughout Indonesia (Susanto et al.,
2020; Wahidiyat et al., 2020; Rujito et al., 2015; Hernanda et al., 2012), while CD41/42 and CD15
found in this study were mutations specific to Cirebon ethnicity.
Table 2 Allele frequency and
genotype distribution for ?-thalassemia gene defects
|
Mutation types |
Frequency |
% | |
|
HBA Alleles |
- |
2 |
3,3% |
|
| - |
58 |
96.7% |
|
HBA Genotypes |
- |
2 |
6.7% |
|
|
|
28 |
93.3% |
Table 3 Allele frequency and
genotype distribution for ?-thalassemia mutations
|
Mutation types |
Frequency |
% | |
|
HBB Alleles |
IVS1-5 |
28 |
46.7% |
|
|
CD26 (HbE) |
19 |
31.7% |
|
|
CD41/42 |
3 |
5% |
|
|
CD15 |
3 |
5% |
|
|
IVS1-1 |
1 |
1.7% |
|
|
IVS1-1 (G>A) |
1 |
1.7% |
|
|
IVS2-654 |
1 |
1.7% |
|
|
Unknown |
4 |
6.7% |
|
HBB Genotypes |
IVS1-5/CD26 |
13 |
43.3% |
|
|
IVS1-5/IVS1-5 |
7 |
23.3% |
|
|
CD41/42/CD26 |
3 |
10% |
|
|
IVS2-654/CD26 |
1 |
3.3% |
|
|
CD15/CD15 |
1 |
3.3% |
|
|
IVS1-5/IVS1-1 |
1 |
3.3% |
|
|
IVS1-1(G>A)/CD26 |
1 |
3.3% |
|
|
CD15/CD26 |
1 |
3.3% |
|
|
Unknown |
2 |
6.7% |
From the HBB alleles, the most common HBB genotypes were IVS1-5/CD26 (43.3%), followed by IVS1-5/IVS1-5 (23.3%), and CD41/42/CD26 (10%), and there were 2 subjects with unknown HBB mutations. This finding can be used to develop a more tailored method for screening and diagnosis, focusing on the most commonly occurring alleles as opposed to rarely occurring alleles (Hassan et al., 2013; Bhardwaj et al., 2005; Varawalla et al., 1991). These four major HBB mutations cover 88.4% of the subjects; developing a quadruplex PCR to screen and diagnose thalassemia can simplify and hasten the diagnostic approach while being cost-effective.
3.3. Genotypic and Phenotypic Correlation
From the HBA and HBB mutation analysis, the
genotypes can be categorized into four main groups (Table 4), with
?-thalassemia/hemoglobin E genotypes as the dominant genotype (60%), followed
by the homozygous ?-thalassemia (26.6%), and two unknown mutations. To
establish the genotype-phenotype relations in transfusion-dependent thalassemia
patients in Cirebon the red blood cell indices between the thalassemia
genotypes were compared unknown excluded (Table 5). The difference between
hemoglobin levels, MCV, MCH, RDW, and hemoglobin A, and disease severity were
not statistically significant between the homozygous thalassemia and
?-thalassemia/hemoglobin E genotypes. In contrast, the HbA2+HbE and HbF levels
between the homozygous
thalassemia and ?-thalassemia/hemoglobin E genotypes
were significant. The
thalassemia/hemoglobin E has a higher HbA2+HbE
percentage (20.28 ± 16.05 vs. 9.91 ± 10.23) compared to their homozygous
thalassemia counterparts. Hemoglobin E is a common structural hemoglobin
variant arising from a mutation in the HBB gene and can be detected with
hemoglobin electrophoresis (Fucharoen &
Weatherall, 2012). This abnormal hemoglobin
is not present in the homozygous ?-thalassemia and can be used as a diagnostic
tool to differentiate between the two genotypes.
Table 4 Thalassemia genotypes of
the transfusion-dependent thalassemia patients
|
Thalassemia genotype |
Frequency |
% |
|
Homozygous ?-thalassemia |
8 |
26.6% |
|
?-thalassemia/CD26 (HbE) |
18 |
60% |
|
Homozygous ?-thalassemia
with ?-thalassemia trait |
2 |
6.7% |
|
Unknown |
2 |
6.7% |
Table 5 Red Blood Indices for
homozygous ?-thalassemia and Blood Indices ?-thalassemia/CD26 (HbE)
|
Parameter |
Homozygous ?-thalassemia (n=8) |
Homozygous
?-thalassemia with ?-thalassemia trait (n=2) |
?-thalassemia/CD26 (HbE) (n=18) |
|
Hb Level (g/dL) |
9.27 ± 2.21 |
10.30 ± 0.42* |
9.32 ± 1.48 |
|
MCV (fL) |
77.30 ± 6.65 |
80.50 ± 2.12* |
73.89 ± 6.82 |
|
MCH (pg) |
24.90 ± 3.14 |
25.50 ± 0.71 |
23.06 ± 2.88 |
|
RDW (%) |
18.21 ± 7.48 |
15.15 ± 3.75 |
20.36 ± 6.55 |
|
HbA |
74.35 ± 10.89 |
80.35 ± 2.62* |
61.81 ± 20.81 |
|
HbA2+HbE (%)+ |
9.91 ± 10.23* |
5.05 ± 3.04* |
20.28 ± 16.05* |
|
HbF (%) |
4.48 ± 3.49* |
2.00 ± 0.42* |
8.72 ± 9.16* |
|
Mahidol Severity Index |
8.0 ± 0.82 |
8.5 ± 0.71 |
7.8 ± 0.77 |
Hb = Hemoglobin, MCV =
Mean Corpuscular Volume, MCH = Mean Corpuscular Hemoglobin, RDW =
Red-cell Distribution Width
+ Hemoglobin A2 and Hemoglobin E on the same value in the HPLC system
(Variant II)
* p
< 0.05
The
HbF levels between the homozygous thalassemia and
thalassemia/hemoglobin E
genotypes were also significant. The HbF level of the
thalassemia/hemoglobin E
is higher than the homozygous ?-thalassemia (8.72 ± 9.16 vs. 4.48 ± 3.49). The
?-thalassemia/hemoglobin E genotype is often associated with higher levels of
HbF and can reduce the severity of the disease. This is due to the results of
XmnI +/+ polymorphism genotype that is often co-inherited with the
?-thalassemia/hemoglobin E genotype (Kesornsit et al., 2018; Rujito et al., 2016). To date, several drugs, such as
thalidomide/lenalidomide, sirolimus, ruxolitinib/pacritinib (JAK2 inhibitors),
and luspatercept/sotatercept (activin receptor-II trap ligands), have been used
to boost the production of HbF in thalassemia patients with hope to alleviate
severity. Hydroxyurea was the first drug to be approved by the FDA and has been
shown to increase the expression of HbF by 2- to 9-fold, while luspatercept, a
recently FDA-approved drug, inhibits over-activated SMAD signaling proteins in
the erythroid precursors (Oikonomidou &
Rivella, 2018; Taher et al., 2018; Soni, 2017).
An
interesting finding in genotype-phenotype was observed in the co-inheritance of
homozygous ?-thalassemia with the ?-thalassemia trait. The RBC indices showed
better parameters and borderline normal indices when compared to the homozygous thalassemia and
thalassemia/hemoglobin E genotypes. It had higher
hemoglobin values, higher MCV and MCH, lower RDW, higher HbA, and lower
HbA2+HbE levels akin to normal phenotype. Co-inheritance with
thalassemia
often improves the phenotypes due to reduced unpaired ?-globin to match the
reduced or absent production of
globin (Taher et al., 2018). However, the alpha trait in this study did not
appear to alleviate the severity, a similar observation that had also been
reported previously (Svasti et al., 2002).
Among
the three groups, the results from this study showed a genotype-to-phenotype
correlation, especially between the homozygous thalassemia and
thalassemia/HbE. This information can be used as a tool for early prediction
of the clinical severity of the disease, assisting in early intervention in
children born with the disease, and promoting prevention through genetic counseling.
Newborn screening for specific alleles of Thalassemia in Indonesia can help
control the number of newborns with thalassemia in the Indonesian population.
The study revealed a genotype and phenotype correlation of the most
common thalassemia mutations in Cirebon, West Java, Indonesia, with four
alleles dominating the genotype, covering 88.4% of the population. A
significant difference in HbA2+HbE and HbF levels was observed between
homozygous thalassemia and
?-thalassemia/HbE. Homozygous
thalassemia with
thalassemia trait has better
hematological parameters compared to homozygous ?-thalassemia and
thalassemia/HbE but does not translate to a better
severity index. The hematological data and clinical appearance are
closely linked to the type of mutation. Molecular analysis can provide a
multitude of information regarding the disease, which can help in the clinical
management of patients, including fine-tuning the blood transfusion
requirements and reducing iron overload. Identification of mutation alleles in
specific regions can be used to predict clinical manifestation and severity
better and develop a more efficient screening and diagnostic method based on
the most common alleles that cover most of the mutations found.
We thank Beben Benyamin and Saovaros Svasti for their
valuable discussions and helpful comments. Mahidol International Postgraduate
Program 2018 and Universitas Swadaya Gunung Jati Internal Research Fund 2019 fundedA this research.
Bhardwaj, U., Zhang, Y.H., Lorey, F.,
McCabe, L.L., McCabe, E.R.B., 2005. Molecular Genetic Confirmatory Testing from
Newborn Screening Samples for The Common African-American, Asian Indian, Southeast
Asian, and Chinese ?-Thalassemia Mutations. American Journal of Hematology,
Volume 78(4), pp. 249–255
Chong,
S.S., Boehm, C.D., Cutting, G.R., Higgs, D.R., 2000a. Simplified Multiplex-PCR Diagnosis
of Common Southeast Asian Deletional Determinants of ?-Thalassemia. Clinical
Chemistry, Volume 46(10), pp. 1692–1695
Chong,
S.S., Boehm, C.D., Higgs, D.R. and Cutting, G.R., 2000b. Single-Tube Multiplex-PCR
Screen for Common Deletional Determinants of ?-Thalassemia. Blood, Volume
95(1), pp. 360–362
Farashi,
S., Harteveld, C.L., 2018. Molecular Basis of ?-Thalassemia. Blood Cells, Molecules, and Diseases, Volume
70(1), pp. 43–53
Fucharoen,
S., Weatherall, D.J., 2012. The Hemoglobin E Thalassemias. Cold Spring
Harbor Perspectives in Medicine, Volume 2(8), p. a011734
Fucharoen,
S., Weatherall, D.J., 2016. Progress Toward the Control and Management of the
Thalassemias. Hematology/Oncology Clinics of North America, Volume 30(2),
pp. 359–371
Giardine,
B., Borg, J., Viennas, E., Pavlidis, C., Moradkhani, K., Joly, P.,
Bartsakoulia, M., Riemer, C., Miller, W., Tzimas, G., Wajcman, H., Hardison,
R.C. Patrinos, G.P., 2014. Updates of the HbVar Database of Human Hemoglobin
Variants and Thalassemia Mutations. Nucleic Acids Research, 42(D1), pp.
1063–1069
Hassan,
S., Ahmad, R., Zakaria, Z., Zulkafli, Z., Abdullah, W.Z., 2013. Detection of
?-Globin Gene Mutations Among ?-Thalassaemia Carriers And Patients In Malaysia:
Application Of Multiplex Amplification Refractory Mutation System-Polymerase
Chain Reaction. The Malaysian Journal of Medical Sciences, Volume 20(1),
pp. 13–20
Hernanda,
P.Y., Tursilowati, L., Arkesteijn, S.G.J., Ugrasena, I.D.G., Larasati, M.C.S.,
Soeatmadji, S.M., Giordano, P.C., Harteveld, C.L., 2012. Towards a Prevention
Program for ?-Thalassemia. the Molecular Spectrum in East Java, Indonesia. Hemoglobin,
Volume 36(1), pp. 1–6
Kesornsit,
A., Jeenduang, N., Horpet, D., Plyduang, T., Nuinoon, M., 2018. Quantitative
Trait Loci Influencing Hb F Levels in Southern Thai Hb E (HBB: c.79G>A)
Heterozygotes. Hemoglobin, Volume
42(1), pp. 23–29
Lawen,
A., Lane, D.J.R., 2012. Mammalian Iron Homeostasis in Health and Disease:
Uptake, Storage, Transport, and Molecular Mechanisms of Action. Antioxidants
& Redox Signaling, Volume 18(18), pp. 2473–2507
Leecharoenkiat,
K., Lithanatudom, P., Sornjai, W., Smith, D.R., 2016. Iron Dysregulation in
Beta-Thalassemia. Asian Pacific Journal of Tropical Medicine, Volume 9(11),
pp. 1035–1043
Malik,
P., Tisdale, J., 2017. Gene and Cell Therapies for Beta-Globinopathies. US:
Springer
Nienhuis,
A.W., Nathan, D.G., 2012. Pathophysiology and Clinical Manifestations of the ?-
Thalassemias. Cold Spring Harbor Perspectives in Medicine, Volume 2(12),
pp. 1–13
Oikonomidou,
P.R., Rivella, S., 2018. What Can We Learn from Ineffective Erythropoiesis in
Thalassemia? Blood Reviews, Volume 32(2), pp. 130–143
Piel,
F.B., Weatherall, D.J., 2014. The ?-Thalassemias. New England Journal of
Medicine, 371(20), pp. 1908–1916
Rujito,
L., Basalamah, M., Mulatsih, S., Sofro, A.S.M., 2015. Molecular Scanning of ?-Thalassemia
in the Southern Region of Central Java, Indonesia; a Step Towards a Local
Prevention Program. Hemoglobin, Volume 39(5), pp. 330–333
Rujito,
L., Basalamah, M., Siswandari, W., Setyono, J., Wulandari, G., Mulatsih, S.,
Sofro, A.S.M., Sadewa, A.H., Sutaryo, S., 2016. Modifying Effect of XmnI,
BCL11A, and HBS1L-MYB on Clinical Appearances: A Study on ?-Thalassemia and Hemoglobin
E/?-Thalassemia Patients in Indonesia. Hematology/Oncology and Stem Cell
Therapy, Volume 9(2), pp. 55–63
Soni,
S., 2017. Novel and Innovative Approaches for Treatment of ?-thalassemia. Pediatric
Hematology Oncology Journal, Volume 2(4), pp. 121–126
Sripichai,
O., Makarasara, W., Munkongdee, T., Kumkhaek, C., Nuchprayoon, I., Chuansumrit,
A., Chuncharunee, S., Chantrakoon, N., Boonmongkol, P., Winichagoon, P.,
Fucharoen, S., 2008. A Scoring System for the Classification of
?-thalassemia/Hb E Disease Severity. American Journal of Hematology, Volume
83(6), pp. 482–484
Susanto,
Z.A., Siswandari, W., Rujito, L., 2020. Cd60 (GTG > GAG)/Hb Cagliari Mutation
Was Found in Scanning of ?-thalassemia Alleles from Patients of East
Kalimantan, Indonesia. Molecular Genetics and Metabolism Reports, Volume
22, p. 100550
Sutcharitchan,
P., Saiki, R., Fucharoen, S., Winichagoon, P., Erlich, H., Embury, S.H., 1995.
Reverse Dot-Blot Detection of Thai Beta-Thalassaemia Mutations. British Journal
of Haematology, Volume 90(4), pp. 809–816
Svasti,
M.L.S., Hieu, T.M., Munkongdee, T., Winichagoon, P., Van-Be, T., Van-Binh, T.,
Fucharoen, S., 2002. Molecular analysis of ?-thalassemia in South Vietnam. American
Journal of Hematology, Volume 71(2), pp. 85–88
Taher,
A.T., Weatherall, D.J., Cappellini, M.D., 2018. Thalassaemia. The Lancet,
Volume 391(10116), pp. 155–167
Thein,
S.L., 2018. Molecular Basis of ? Thalassemia and Potential Therapeutic Targets.
Blood Cells, Molecules, and Diseases, Volume 70, pp. 54–65
Varawalla,
N.Y., Old, J.M., Sarkar, R., Venkatesan, R., Weatherall, D.J., 1991. The Spectrum
of ??thalassaemia Mutations on the Indian Subcontinent: the Basis for Prenatal
Diagnosis. British Journal of
Haematology, Volume 78(2), pp. 242–247
Viprakasit,
V., Ekwattanakit, S., 2018. Clinical Classification, Screening and Diagnosis
for Thalassemia. Hematology/Oncology Clinics, Volume 32(2), pp. 193–211
Wahidiyat,
P.A., Sari, T.T., Rahmartani, L.D., Setianingsih, I., Iskandar, S.D.,
Pratanata, A.M., Yapiy, I., Yosia, M., Tricta, F., 2020. An Insight Into Indonesian
Current Thalassaemia Care And Challenges. ISBT Science Series, Volume 15(3),
pp. 334–341
